Targeted regulation of endogenous genes to achieve analysis of target gene expression networks has always been a hot spot in the field of plant research. With the development of CRISPR/Cas technology, the RNA-guided CRISPR activation (CRISPRa) system has been developed and applied in animals and plants. The fusion of dCas9 nuclease and transcription activator can regulate the transcription of endogenous genes and realize the directional upregulation of genes. Precise upregulation of gene transcription is becoming an effective solution for functional genomics research, molecular breeding, and germplasm innovation. However, there are no reports on the application of the CRISPR/dCas9 transcriptional activation system in cotton.
On April 13, 2023, Plant Communications published online a paper entitled "Developing an efficient CRISPR/dCas9-TV derived transcriptional activation system to create three novel cotton germplasm materials" co-authored by Huazhong Agricultural University and Institute of Cotton Research of Chinese Academy of Agricultural Science. In this paper, a transcriptional activation (gene knockout) system based on the CRISPR/dCas9 system was used to regulate endogenous genes to achieve germplasm innovation in cotton.
Based on the optimized CRISPR/dCas9-TV system, the researchers achieved efficient and robust upregulation of cotton endogenous genes GhTULP34 and GhTLP19. The expression of GhTULP34 was upregulated up to 35.5 times and that of GhTLP19 was upregulated up to 30.5 times. Judging from the activation efficiency of GhTLP19 (sgRNA1~10) and GhTULP34 (sgRNA11~20), most sgRNAs can guide dCas9-TV to activate the reporter gene to varying degrees, but the target sites of sgRNAs have a significant impact on the efficiency of transcriptional activation. In order to test whether the up-regulation of GhTULP34 could inhibit stomatal closure when the plants were subjected to osmotic stress, after the transgenic lines were treated with 400mM mannitol, the number of open stomata of TULP#5 plants was significantly more than that of the control, and the directional up-regulation of GhTULP34 improved the drought resistance of cotton.
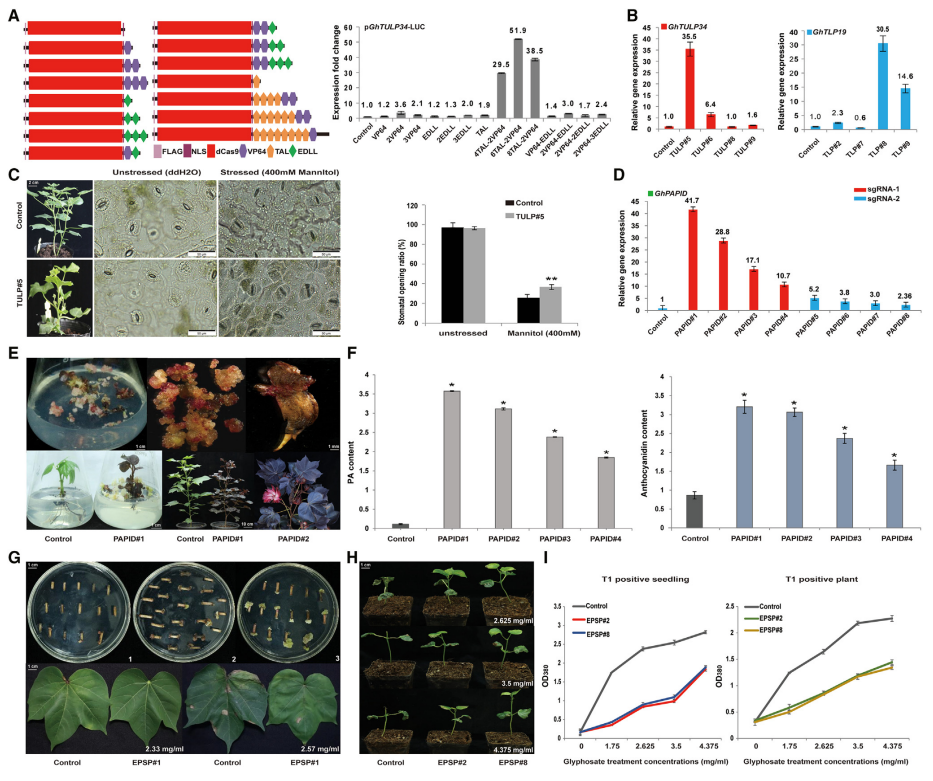
Under the guidance of creating high-quality germplasm resources, the researchers used this system to successfully activate the gene GhPAPID, and its expression was up-regulated by up to 41.7 times. Interestingly, the GhPAPID-activated material showed obvious red/purple phenotype in the callus during plant tissue culture, and this gene can be used as a good reporter gene in the process of genetic transformation of cotton. In addition, leaves, stems, branches and petals of transgenic plants with this gene exhibited a distinct red/purple color. Proanthocyanidin and anthocyanidin contents were significantly increased in different transgenic lines, a result consistent with increased transcript levels in these plants. This research offers an interesting direction for creating naturally colored cotton. In the future, this system may be used in combination with tissue-specific promoters to simultaneously target and up-regulate multiple genes in the flavonoid pathway (such as GhTT2 and GhCHS) to achieve changes in cotton color.
Herbicide resistance is a major functional characteristic of transgenic crops, and breeding herbicide-resistant crops has important economic benefits and obvious environmental benefits. The researchers activated the endogenous GhEPSPS gene in cotton through this system, and successfully obtained transgenic plants whose expression levels were increased by 2.9 to 33.5 times. Glyphosate treatment of the transgenic T0 generation plants found that the transgenic lines showed good glyphosate resistance, and the resistance could be stably inherited from the T0 parent plants to the T1 generation.
Altogether, this study demonstrates that the CRISPR/dCas9-TV system is an efficient gene "knock-up" strategy. Based on the successful activation and creation of three cotton germplasms (coloured, glyphosate-resistant and drought-tolerant germplasms) based on this system, it provides a new research strategy for molecular breeding of tolerant germplasms and innovation of germplasm resources.
Reference:
Yu, L., Li, Z., Ding, X. et al. Developing an efficient CRISPR-dCas9-TV-derived transcriptional activation system to create three novel cotton germplasm materials. Plant Commun. 2023, 4(4): 100600.