More than 70% of the world's crops are polyploid, including wheat, cotton, rapeseed, peanuts and other crops. In addition, corn and soybeans are paleopolyploids, and crops such as rice are partial polyploids. Therefore, polyploidy research has important scientific significance and breeding value for crop genetic improvement.
Compared with diploid ancestral species, polyploid species have obvious advantages in both vegetative and reproductive growth. This advantage is called polyploid heterosis, polyploid advantage, or polyploid vigor. Polyploidy advantage is a kind of fixed hybrid vigor. Further development and utilization of polyploidy advantage will likely promote breakthrough progress in polyploid crop improvement. However, for a long time, the analysis of polyploid crops has been based on the analysis methods of diploid crops, and the research, development and utilization of the advantages of polyploidy have seriously lagged behind.
On October 28, 2023, Molecular Plant published online the collaborative research paper "Genome resources for the elite bread wheat cultivar Aikang 58 and mining of elite homeologous haplotypes for accelerating wheat improvement" by Henan Agricultural University, the Institute of Crop Science of the Chinese Academy of Agricultural Sciences, and the Henan Institute of Science and Technology. The researchers used the elite wheat cultivar Aikang 58 (AK58) as material to establish a multi-omics database of this variety. The analysis found that diploidization and divergence among the A, B, and D subgenomes of wheat are the genomic basis for polyploidy advantage. On this basis, a novel polyploid crop association analysis method: homoeologous locus-based GWAS (HGWAS) was proposed. This method was used to detect and analyze more than 20 important agronomic traits, and a series of important discoveries were made.
Sequencing, Assembly and Database Construction of AK58
The reference genome contig N50 is 237.2 Kb, the scaffold N50 reaches 715 Mb, and the final genome size is 14.75 Gb. Genome annotation analysis obtained 119,448 high-confidence protein-coding genes. AK58 genome comprehensive database (multi-omics database) has been established: including genes, transposable elements (TE), transcriptome, epigenome, metabolome, mutants, genome-based SNPs, QTLs and genetic maps, etc., providing rich multi-omics information for wheat and other related research.
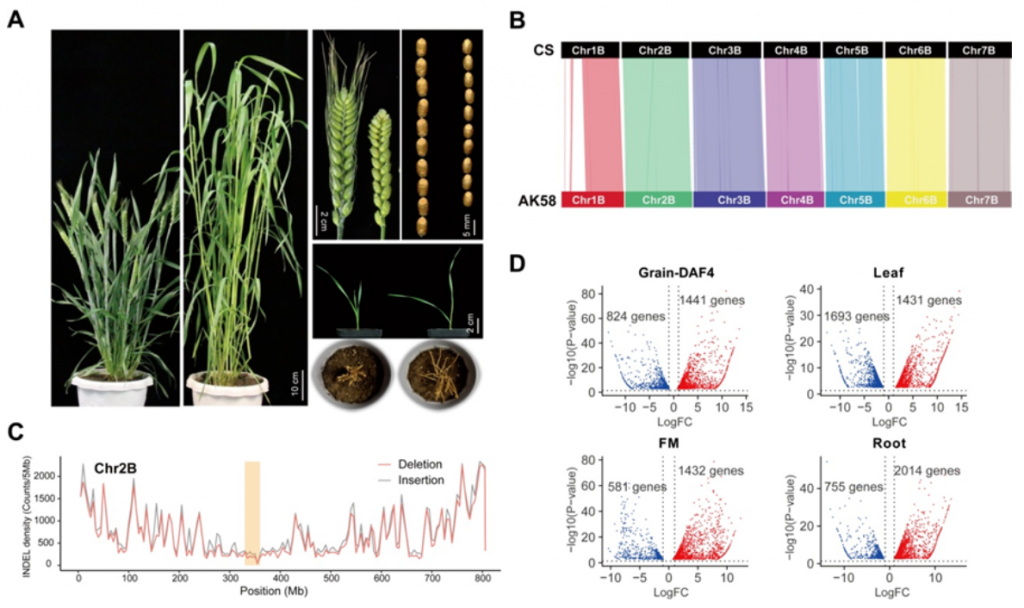
Comparative Genomics Between AK58 and Chinese Spring (CS) Reveal Genomic Changes Induced By Modern Wheat Breeding Improvements
CS is a classic landrace cultivar, and AK58 is a typical modern cultivar, with obvious differences in phenotype. Comparison of the two genomes revealed genome-level variations caused by improvements in wheat varieties over the past century. The genomes of the two varieties have a collinear relationship of 86% - 88%; there are 40.6M (million) SNPs and 5.5M Indels; the specific genes of AK58 and CS are 5857 and 3080 respectively. Genes specifically expressed by AK58 were significantly enriched in photosynthetic system I, damage response, and oxidative phosphorylation pathways, while genes specifically expressed by CS were enriched in plant-pathogen interaction pathways. Transcriptome analysis found that the number of up-regulated genes in AK58 was higher.
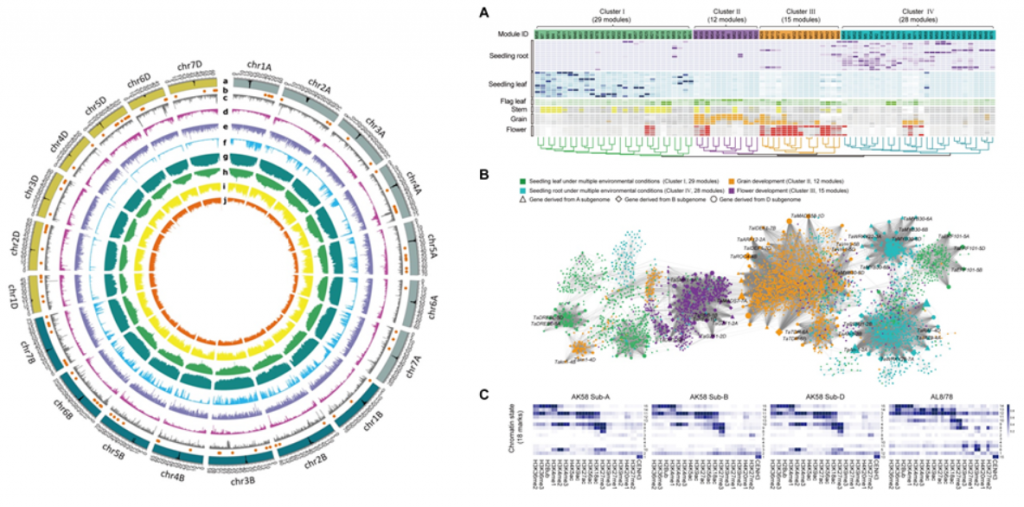
Subgenomic Diploidization and Differentiation, HGWAS and Modern Breeding Improvement
The study found that in hexaploid wheat, three subgenomes undergo simultaneous diploidization and differentiation. Diploidization allows different subgenomes to coordinate their functions; differentiation allows them to generate diversity. The two work together to form a fixed hybrid from polyploidy. Three subgenomic orthologs, or homoeologs can produce a huge number of homoeoallele haplotype (HH), and the HHs multiply exponentially. For example, in an F2 genetic population, as many as 27 orthologous genes can occur between the allelic subgenomes of the two parents. In natural populations, the number is even larger. Therefore, from the perspective of HH, polyploidy such as wheat has extremely high diversity. It is of great significance to exploit the diversity of some homologous alleles. To this end, the authors developed HGWAS. This method was used to detect 20 important agronomic traits of the F2 segregation population composed of AK58 and CS. A total of 393 significant HGWAS loci were found, which was much higher than the 241 loci detected by conventional GWAS, indicating that this method is an effective method for detecting polyploid important agronomic trait loci. Comparing the HH composition of these loci, not only improved HHs from modern breeding were detected, but also HHs with important potential value in future wheat improvement were discovered.
The authors believe that HGWAS loci reflect the comprehensive (interaction) genetic effects between orthologous genes in multiple subgenomes. In polyploid crops, the identification of HGWAS loci, especially the excavation and utilization of excellent HHs, will greatly enrich the genetic diversity of polyploid loci, and will be of great importance and value for further analyzing and utilizing the advantages of polyploidy and cultivating more and better new varieties in the future.
Reference:
Jia, J., et al. Genome resources for the elite bread wheat cultivar Aikang 58 and mining of elite homeologous haplotypes for accelerating wheat improvement. Mol Plant. 2023, 16(12): 1893-1910.