Protein phosphorylation plays an important role in plant growth, development, environmental adaptation, and crop yield and quality regulation. Deep analysis of the phosphoproteome is an effective means to comprehensively understand how phosphorylation functions. However, in-depth analysis of plant phosphoproteomes is technically more challenging compared to animals. Because plant cells have dense cell walls and a large amount of secondary metabolites including pigments, the former greatly increases the difficulty of protein extraction, while the latter seriously interferes with the efficiency and specificity of phosphopeptide enrichment.
On November 27, 2023, Wang Yingchun's team from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences published a research paper titled "GreenPhos, a universal method for in-depth measurement of plant phosphoproteomes with high quantitative reproducibility" online in Molecular Plant. The paper reports a breakthrough new technology for plant phosphoproteomics. This technology adopts a simplified and robust workflow, effectively overcoming the main technical difficulties in plant phosphoproteome analysis, and can quickly enrich plant phosphopeptides with high sensitivity and specificity. This technology can be used to quantitatively analyze the phosphoproteomes of different plants. Its analysis depth and quantitative reproducibility are unprecedented, and it is expected to become a universal technology for plant phosphoproteomics. Because this technology is mainly aimed at higher plants and other green organisms (such as Chlamydomonas), and is easy to operate, it greatly reduces the labor and reagent costs required for experiments, so it is named GreenPhos.
Researchers have applied this method combined with mass spectrometry technology to analyze the phosphoproteomes of a variety of green organisms. A single mass spectrometry analysis can identify approximately 11,000 phosphate sites, reaching an internationally leading level. The main feature of this technology is the efficient extraction of plant proteins using sodium deoxycholate, while eliminating two steps in conventional methods that lead to sample loss and reduced sensitivity, that is, sample purification before protease digestion and desalting before phosphopeptide enrichment, high-specificity and high-sensitivity enrichment of phosphopeptides in the presence of pigments and other interfering molecules. Due to minimal sample loss during the operation, the sensitivity, quantitative reproducibility, and time cost-effectiveness of phosphoproteome analysis are greatly improved, while experimental errors are effectively reduced. This technology has been used to deeply analyze the differential phosphoproteome of Arabidopsis thaliana in response to different durations of salt stress, and discovered phosphorylation events including spliceosome proteins and some kinases in response to salt stress.
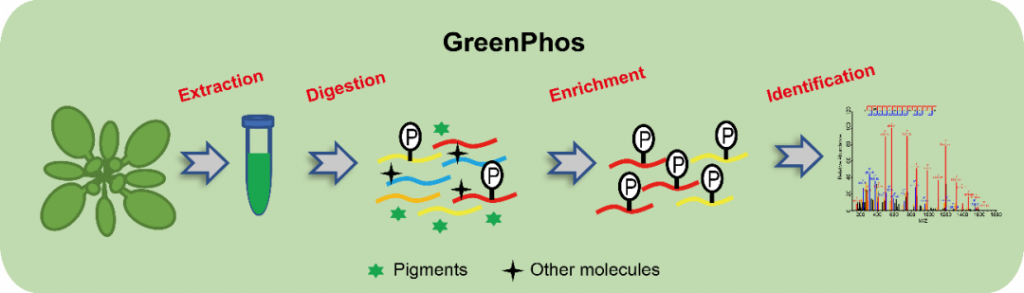
GreenPhos not only greatly improves the analysis efficiency of plant phosphoproteome, but also significantly reduces the difficulty and cost of experimental operations, providing a powerful tool for a deeper understanding of the function of protein phosphorylation in plant life processes. The research results will effectively promote the cross-integration of phosphoproteomics with fields such as plant biology and agronomy, and have broad application prospects in discovering phosphorylated proteins and their sites that are closely related to crop yield, quality and stress resistance.
Reference:
Duan, X., et al. GreenPhos, a universal method for in-depth measurement of plant phosphoproteomes with high quantitative reproducibility. Mol Plant. 2023, Nov 27: S1674-2052(23)00393-3.