Regulatory elements are key components of plant genomes, shaping the domestication and improvement of modern crops. However, their identity, function, and diversity remain poorly understood, limiting our ability to exploit their agricultural potential in natural or induced variation.
On June 12, 2025, Andrea Gallavotti's team at Rutgers University and his collaborators published a paper titled "Transcription factor binding divergence drives transcriptional and phenotypic variation in maize" in Nature Plants.
This study mapped the binding of 200 transcription factors (TFs) from 30 families in two commonly used inbred lines for maize breeding (B73 and Mo17). TF binding comparisons revealed extensive differences between inbred lines, mainly driven by structural variations (SVs), associated with changes in gene expression, and explained complex quantitative trait loci (such as the flowering time regulator Vgt1 and the insect resistance enhancer DICE). By editing TF binding regions with CRISPR-Cas9, the functions of loci regulating plant architecture and bioresistance were verified. The maize TF binding catalog of this study identified functional regulatory regions, supported integration and comparative analysis across genotypes, and provided important resources for agricultural improvement.
Technology: DNA affinity purification sequencing (DAP-seq) was used to analyze the genomic binding sites of 200 TFs in B73 and Mo17, and 400 high-quality data sets were obtained.
Coverage: Thousands of binding events were identified, covering cis-regulatory regions such as promoters and enhancers.
Subfamily specificity: Members of different branches of the same TF family (e.g., subfamilies of bZIP) have unique binding preferences.
Functional association: GO analysis shows that TF target genes are enriched in specific pathways (e.g., HSF24 regulates heat stress response).
Agronomic trait association: TF binding sites are significantly associated with GWAS trait SNPs (e.g., GRF/bZIP is associated with leaf angle trait).
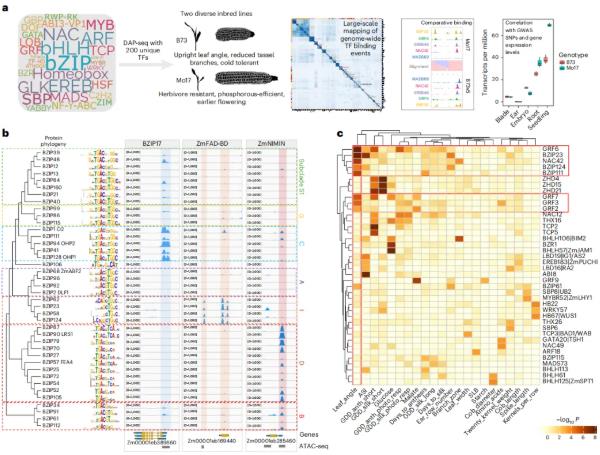
Figure 1. Large-scale DAP-seq profiling of maize TFs provides high-quality genome-wide binding site data for B73v5 and Mo17.. (Galli, et al., 2025)
DAP-CRMs identification: Development of a TF diversity panel set (66 representative TFs) to identify 225,235 cis-regulatory modules (DAP-CRMs), with an average of 5 TF binding sites.
Functional validation: DAP-CRMs highly overlap with chromatin open regions (ATAC-seq) and conserved non-coding sequences, and locate known QTLs (e.g., GRASSY TILLERS1).
Combinatorial regulatory effects: Specific TF combinations (e.g., ARF + SBP + NAC) synergistically regulate traits (e.g., leaf angle vs. tillering).
Flowering time regulation: MITE insertion in the Vgt1 enhancer in Mo17 disrupts MADS69 binding sites, leading to early flowering.
ARF binding variation: 12-bp insertion in B73 forms TGTC repeats, enhancing ARF binding.
Structural variation (SV) dominance: 37% of TF binding sites are genotype-specific, with SV being the main driver.
Classic case
Position variation (PosV): 17% of conserved binding sites undergo >500 bp displacement, affecting target gene expression (e.g., ZmATL34 expression increased 13-fold).
New mechanism discovery: Gln-Ala-Arg-type bHLH (VIII subclass) binds DNA through monomers and heterodimers (with ZmSPT), recognizing the non-classical motif CCCATACC.
Technical innovation: Development of doubleDAP-seq to analyze dimer binding preference, and the binding site is significantly different from that of the monomer.
TSH1 promoter: Deletion of DAP-CRM containing SBP30 leads to ectopic growth of bracts.
DICE enhancer: Deletion of tandemly duplicated TF binding sites (including ARF/GLK/BZIP) significantly reduces the expression of insect resistance gene BX1.
Driving mechanism of regulatory variation: SV is the main source of TF binding differences between maize inbred lines, directly related to gene expression and phenotypic diversity (such as flowering time, insect resistance).
Importance of coordinated regulation: TFs form functional modules (DAP-CRMs) by clustered binding, and specific combinations precisely regulate complex traits.
Evolution and conservation: 63% of TF binding events are conserved between inbred lines, and the core regulatory network is stable in evolution; the remaining variation provides the basis for phenotypic plasticity.
Application potential: The regulatory map provides targets for designing alleles (such as CRISPR editing enhancers), which helps precision breeding.
The first maize genome-wide TF binding map: covering 200 TF binding sites in two genotypes, and establishing open resources (GSE275897).
Revealing the core role of SV in regulatory variation: breaking through the SNP-centric paradigm, clarifying that structural variation drives phenotypic adaptation by disrupting TF binding.
Development of doubleDAP-seq technology: analyzing heterodimer binding specificity and discovering new DNA binding mechanisms of plant-specific bHLH subclasses.