I often hear people around me arguing from time to time: "Is genetically modified food safe?" "What are the pros and cons of genetically modified technology?" and other issues. People always want to avoid modern products as much as possible, pursue returning to nature, and regard genetic modification as an act that interferes with nature. However, our food and clothing are inseparable from the products of genes, so starting with genes seems to be the best choice.
Nature also seems to be giving us hints. On October 5, 2023, the team of Luke T. Dunning at the University of Sheffield in the United Kingdom published a research paper titled "Lateral gene transfer generates accessory genes that accumulate at different rates within a grass lineage" in the New Phytologist magazine. The study used whole-genome sequencing and bioinformatics analysis to find that grasses can incorporate DNA from other species into their genomes through lateral gene transfer (LGT), giving them evolutionary advantages such as faster growth and greater adaptability. This is further evidence that grasses do not evolve according to rules, but rather steal beneficial genes from their neighbors through LGT to outperform other species.
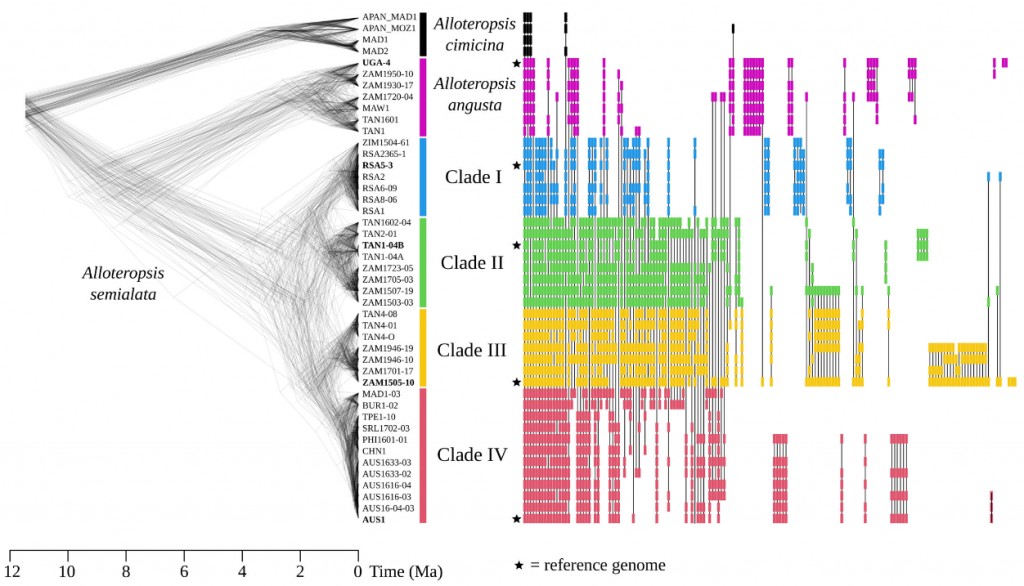
This study used five newly assembled genomes of the grass species (Alloteropsis) as research objects, including three A. semialata (R. Br.) Hitchc. Genomes and the previously obtained sequenced genome of Australian origin species. These four genomes cover the four main nuclear subgroups of this species. In addition, the study also obtained the genome of A. angusta Stapf from the Ugandan source species (accession AANG_UGA4).
These different genomes provide genomic information of different nuclear subgroups within Alloteropsis species, which is helpful for studying various biological and evolutionary issues and plays a key role in deeply understanding the evolution and diversity of Alloteropsis genomes.
Through phylogenetic tree analysis, the authors identified a total of 168 laterally acquired genes. These genes came from three different subfamilies: 92.2% from Panicoideae, 4.8% from Chloridoideae, and 3.0% from Danthonioideae. Within Panicoideae, the main donors are Cenchrinae (88 LGTs), Andropogoneae (54) and Melinidinae (7).
Although the stochastic mapping method is stochastic in molecular clock analysis, the origin age inferences derived from 100 repeated experiments vary. However, the analysis results show a consistent trend, that is, the observed lateral acquisition events have increased significantly in the recent past, especially in the ZAM1505-10 genome. When calculating the average of pseudo-replicate experiments, it was observed that the number of transversely acquired genes increased significantly in the recent past in RSA5, AUS1, and ZAM1505-10, with an exponential distribution in the ZAM1505-10 genome. According to the fitted exponential decay model, the rate of lateral acquisition of genes in the five genomes ranges from approximately 2.66 to 15.74 LGT per Ma. The highest value occurs in ZAM1505-10, whose rate of increase is significantly higher than that of other genomes. The rate of lateral acquisition of genes lost per Ma in A. angusta is estimated to be 4%, consistent with a recent slight increase in lateral acquisition events. Modeling analyzes indicate that 11% to 16% of laterally acquired genes were lost per Ma in most A. semialata lineages, and 24% were lost in the lineage of ZAM1505-10, although the confidence intervals for the latter overlap with those of some other lineages. Therefore, the large number of lateral gains in the ZAM1505-10 genome is due to a higher rate of growth, although there is also a higher rate of loss. Taken together, these analyzes indicate that the Alloteropsis genome undergoes a high frequency of lateral acquisition of genes, with lateral acquisition events occurring throughout its evolutionary history followed by relatively rapid loss of these genes. However, the rates of gain and loss vary between different sublineages, making the A. semialata genome harbor particularly important variation in lateral gene acquisition.
In this study, the authors focused on five reference genomes to assess pan-genome variation. Of the 168 laterally acquired genes, only 2 are present in the five A. semialata and A. angusta reference genomes and thus can be considered core LGT genes, while the vast majority (166 genes, 98.8%) are considered accessory LGT genes.
In this study, the authors focused on five reference genomes to assess pan-genome variation. Among the 168 laterally acquired genes, only 2 are present in the five A. semialata and A. angusta reference genomes and therefore can be considered core LGT genes, while the vast majority are considered accessory LGT genes. Of the 166 polymorphic laterally acquired genes, 21 are unique to A. angusta and 136 to A. semialata, with the other 9 shared in some genomes of both populations. Of the genomic positions unique to A. semialata, 130 have undergone internal sequence variation in A. semialata, while most of the genome-wide variation is caused by later lateral acquisition of genes that are only present in ZAM1505 and, to a lesser extent, the AUS1 genome. These results indicate that recent lateral acquisition events and subsequent gene losses resulted in important pan-genomic variation within A. semialata and between the two sister species. Unlike genes acquired through lateral gene acquisition, only 3.4% of the 6657 native genes were considered accessory genes, which was mainly caused by the loss of many genes in the A. angusta lineage and thus accounted for a larger proportion of the native pan-genome. In the A. semialata genome, the four source species contributed to local pan-genome variation in similar proportions, which is in sharp contrast to the pan-genome contribution of later lateral acquired genes.
Gramineae have important ecological and economic value. The research team sequenced multiple genomes of a tropical grass and believed that these gene transfers are likely to be the same or similar to the way some genetically modified crops are produced. Therefore, this process can be used to increase crop yields and produce more stress-resistant crops, while allowing people to view and use controversial genetically modified crops more rationally.
As Dr. Luke Dunning said, there are many ways to create genetically modified crops, some require a lot of human intervention, some do not, and some of them will occur naturally with minimal human intervention, so the results of gene transfer will be observed in wild species.
This means that in the near future, the controversial genetic modification may be regarded as a more natural process.
Reference:
Raimondeau, P., Bianconi, M.E., Pereira, L. et al. Lateral gene transfer generates accessory genes that accumulate at different rates within a grass lineage. New Phytol. 2023, 240(5): 2072-2084.