On August 14, Nature Communications published online a research paper entitled "Identification of gene function based on models capturing natural variability of Arabidopsis thaliana lipid metabolism" by the teams of Zoran Nikoloski and Sandra Correa Córdoba from the University of Potsdam in Germany, Max Planck Institute of Molecular Plant Physiology and their collaborators. This study builds a Plant Lipid Module (PLM), which can be easily integrated into existing metabolic models. And through three case studies, the application and predictive accuracy of PLM are demonstrated, providing a basis for detailed research on the regulation of lipid composition.
Lipids consist of a class of structurally diverse molecules that facilitate compartmentalization of biochemical activities and participate in key cellular functions. The structural diversity of plant lipids is generated by the underlying metabolic network, and fine-tuning the lipidome can better adapt plants to internal and external environments, playing an important role in regulating agronomically important traits. At the same time, plant lipids are closely related to agricultural and biotechnological applications. Currently, advances in plant lipid metabolism are still largely based on reductionist approaches. In addition, through genome-scale models (GEMs), lipid metabolism can be mechanistically described, such as characterizing how lipid metabolism is interconnected with primary metabolic pathways, or designing manipulation strategies for lipid-related traits. However, existing models of plant lipid metabolism can only provide a focused representation, severely hampering detailed studies of the regulation of specific components.
In order to solve the above problems, this study established the PLM, which contains 5956 biochemical reactions and 3108 metabolites in 16 compartments. Alternatively, PLM can be directly integrated into existing metabolic models, including five published plant metabolic models, through easy-to-use tools.
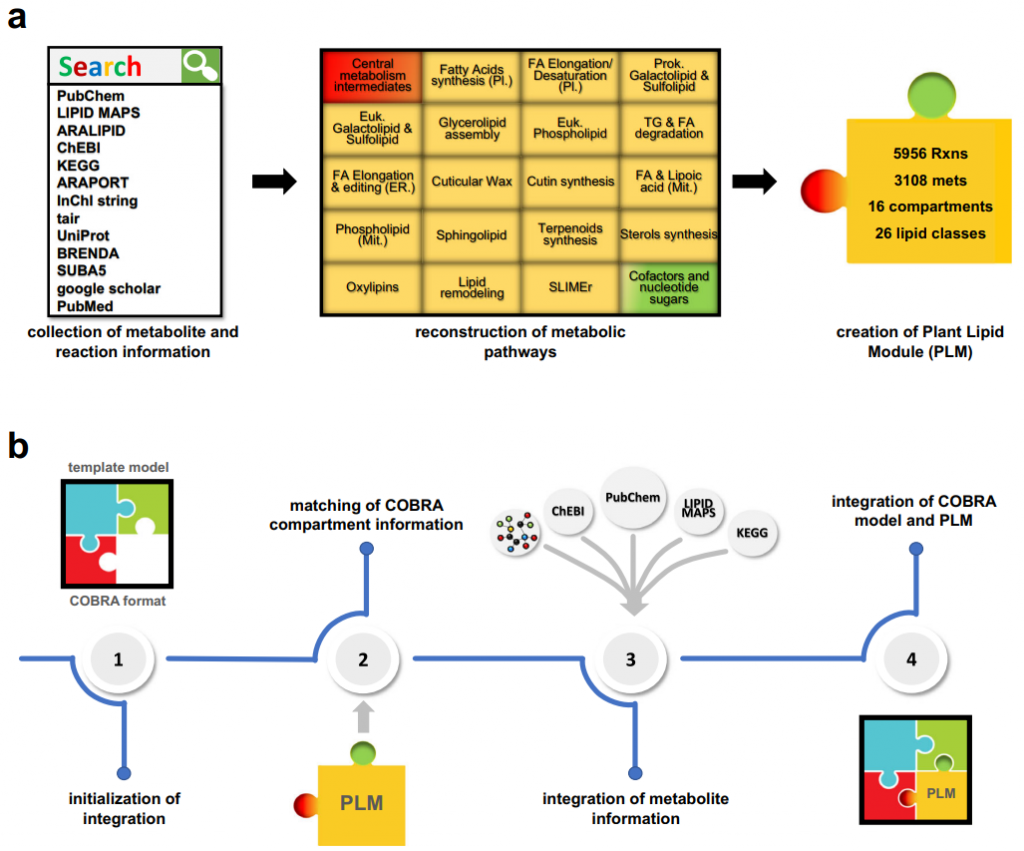
By integrating into the metabolic model of Arabidopsis Col-0, this study shows that PLM can generate accurate predictions (83%) for single lethal knock-outs. These metabolic pathways include the production of lipids, carbohydrates, nucleotide sugars, ATP, and sugar phosphates. In addition, PLM can be integrated with accession-specific metabolic models and provide a basis for studying natural variation and genetic architecture of lipid metabolic fluxes.
Next, the researchers exploited the response activation/inactivation patterns and the corresponding increase/decrease in gene expression in Arabidopsis thaliana exposed to prolonged darkness. The study found that transcripts measured in the dark correlated with predicted flux changes with 75% agreement. Although the researchers speculated that the remaining 25% may be related to post-translational modifications (PTMs); however, the analysis did not reveal significant differences in the types and proportions of PTMs between concordant enzymes and outlier enzymes.
Through genome-wide associations with fluxes obtained by integrating PLMs into diurnal conditions and accession-specific models, this study identified up to 65 candidate genes regulating lipid metabolism in Arabidopsis thaliana. Using published mutants, the study validated 40% of these candidate genes, paving the way for identifying the function of metabolic genes based on models that capture natural variation in metabolism.
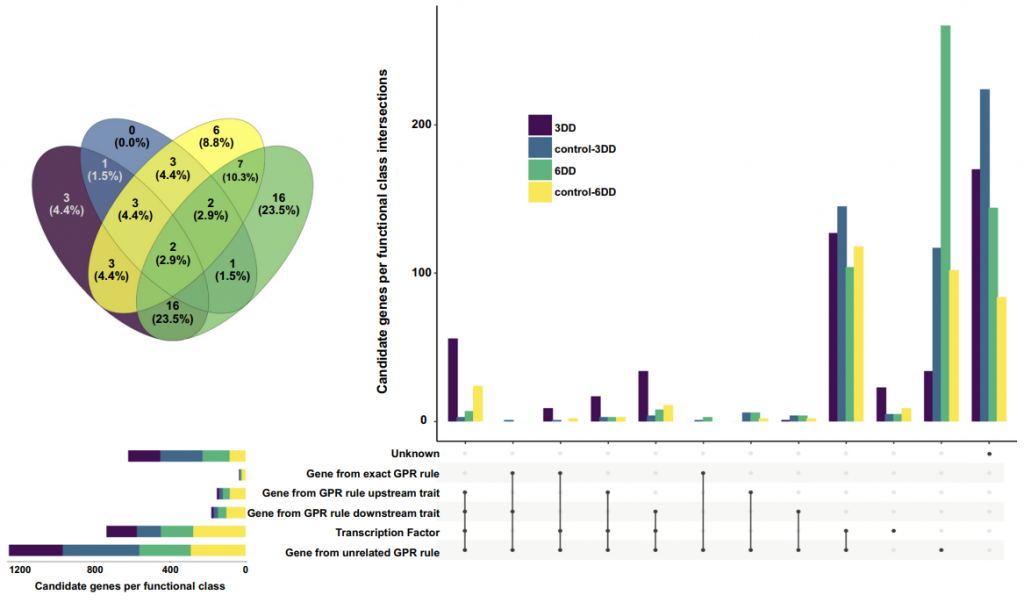
Taken together, this study establishes a PLM that can be easily integrated into metabolic models of any scale in a semi-automatic manner. The application and predictive accuracy of PLM are demonstrated through three case studies integrating PLM into five plant metabolic models: gene lethality, lipid metabolism in response to prolonged darkness, and putative lipid metabolism candidate genes. Thus, PLM predictions provide new insights into the model-driven natural variability in lipid metabolism in Arabidopsis germplasm, demonstrating its utility as an experimental genome-wide association study.
Reference:
Córdoba, S.C., Tong, H., Burgos, A. et al. Identification of gene function based on models capturing natural variability of Arabidopsis thaliana lipid metabolism. Nat Commun 14, 4897 (2023).