Endoplasmic Reticulum Associated Degradation for ER Stress in Plants
The endoplasmic reticulum-associated degradation (ERAD) machinery clears the misfolded protein substrate through the cytoplasmic 26S proteasome. In plants, ERAD not only plays an important role in normal growth and development but also is related to the process of tolerance to stress. The process consists of four steps, recognition of misfolded proteins, inversion of proteins located in the ER lumen and ER membrane to the cytoplasm, ubiquitination modification under the action of specific E1, E2, and E3 ligases, and degradation by 26S proteasome.
Lifeasible makes full use of our specialized libraries, selection strategies, and maturation techniques to analyze the four steps in the process of endoplasmic reticulum-associated degradation. With advanced technology and experienced staff, we provide comprehensive ER stress response services for global clients to support their research.
Recognition of Misfolded Proteins
- The recognition of misfolded proteins mainly involves two ways; one is the recognition of misfolded glycoproteins, which mainly depends on the α-1, 6 glycosidic bonds exposed after the glycoprotein is cleaved. The other is the recognition of non-glycoproteins, accomplished by BiP recognition of hydrophobic peptides exposed by misfolded proteins.
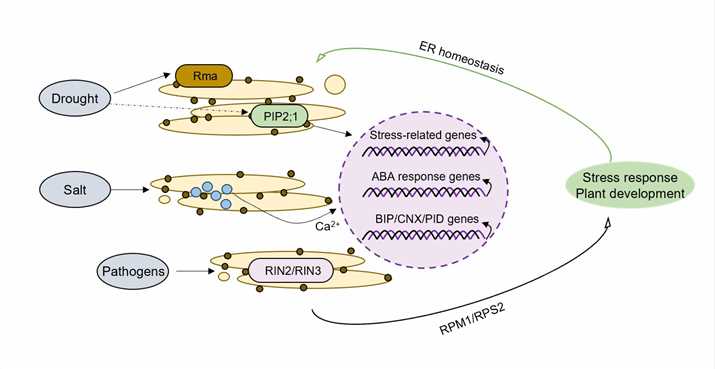 Fig.1. Endoplasmic reticulum-associated degradation plays crucial roles in biotic and abiotic stress responses.
Fig.1. Endoplasmic reticulum-associated degradation plays crucial roles in biotic and abiotic stress responses.
- Lifeasible provides services included in the biological process of identifying misfolded glycoproteins. OS9 in plants interacts with HRD3A after binding the α-1, 6 glycosidic bonds to pull the substrate into the HRD1 complex.
- We also provide services to identify non-glycoprotein biological processes. The protein binds to BiP and is passed to disulfide reductase, which then enters the ERAD system.
Inversion of Proteins from the ER to the Cytoplasm
- Most erroneous proteins removed by the ERAD system are endoplasmic reticulum lumen or membrane proteins, which need to be transported to the cytoplasm before they can be degraded by the cytoplasmic proteasome.
- Lifeasible helps our customers study this process through three anti-transport channels, such as the SEC61 transport channel, DER1 transport channel, and E3 ligase that mediates substrate protein ubiquitination modification.
Ubiquitination Modification
- Ubiquitination of the substrate is carried out after inversion to the cytoplasm.
- In plants, we help to study the key enzymes in ubiquitination modification, including E3 ligases HRD1, DOA10, and Rma1, adaptor protein HRD3A, ubiquitin-binding enzyme UBC32, etc.
Lifeasible offers professional services covering endoplasmic reticulum-associated degradation to meet your research needs. With years of experience in plant sciences, our advanced platforms can help our clients solve various difficulties and conduct research. If you are interested in our services or have any questions, please feel free to contact us or make an online inquiry.
For research or industrial raw materials, not for personal medical use!
 Fig.1. Endoplasmic reticulum-associated degradation plays crucial roles in biotic and abiotic stress responses.
Fig.1. Endoplasmic reticulum-associated degradation plays crucial roles in biotic and abiotic stress responses.