On August 10, 2023, Nature Plants published online a research paper titled "A multi-omic Nicotiana benthamiana resource for fundamental research and biotechnology" by the joint team of Peter M. Waterhouse and Jiyuan An of Queensland University of Technology, Christopher Winefield of Lincoln University, and their collaborators. This study constructed and analyzed the whole genome, epigenome, metabolome, and single nucleotide polymorphism (SNP) map of the N. benthamiana LAB strain and wild strain QLD, providing an important basis for further application of N. benthamiana in scientific research and biofactory.
N. benthamiana is an important model plant with an allotetraploid genome of approximately 3 Gb. At the same time, N. benthamianais a very important plant platform for biopharmaceutical protein and vaccine production, and has played an important role in research discoveries in RNA interference (RNAi), plant-pathogen interactions, metabolic pathway engineering, functional genomics, synthetic biology and gene editing. Therefore, N. benthamiana is an important LAB plant. However, compared with Arabidopsis thaliana, its various omics data are still lacking, severely limiting its practical application.
To further improve the practicality and versatility of N. benthamiana, this study performed high-quality, chromosome-level genome assembly of commonly used LAB strains and wild strain QLD, and the construction of transcriptome, epigenome, microRNA and transposable element databases. In addition, the study further constructed SNP maps for two laboratory strains and four wild strains.
Using dendrogram analysis of matrices of degrees of similarity of counterpart gene sequences of the Nicotiana set, the researchers clearly identified 8 homeologous chromosome pairs and 3 orphan chromosomes. Although features such as the loss of 5 chromosomes in the ancestral tetraploid, enlarged intergenic regions, and extensive segmental allopolyploidy have not been found in other tobacco genomes, the two subgenomes in N. benthamiana show extensive synteny throughout the Solanaceae family. Subgenomes A and B encode 23,408 and 22,388 genes, respectively, and the overall transcript abundance of homologous genes differs by only 1%.
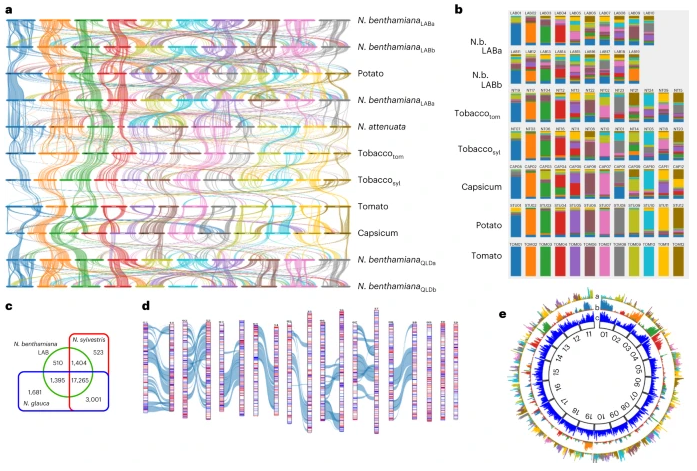
Impairment of RNAi response mechanisms in LAB strains may underlie their use as biofactory and research tools. This study found that there are many genetic, metabolic and phenotypic differences between LAB and QLD strains, including different RNAi responses. But at the same time, they are highly interfertile and both are suitable for genome editing, as well as transient or stable transformation.
Finally, the study analyzed transposons, epigenetic profiles and gene density in N. benthamiana. Due to the loss of 5 chromosomes in the ancestral allotetraploid, approximately 50% of the genes in the N. benthamiana genome remain as singletons. Interestingly, N. benthamiana chromosomes, especially chromosomes 5-10 and 15-19, have a more uniform gene density. This unusual arrangement may be due to their formation through abundant interchromosomal recombination.
In summary, this study released the full genome, epigenome, and metabolome information of LAB strains and QLD strains, as well as their SNP maps. The establishment of these big data resources is expected to make the N. benthamiana LAB/QLD combination as practical as the Col-0/Ler of A. thaliana, further promoting the application of N. benthamiana in scientific research and biofactory.
Reference:
Ranawaka, B., An, J., Lorenc, M.T. et al. A multi-omic Nicotiana benthamiana resource for fundamental research and biotechnology. Nat. Plants 9, 1558–1571 (2023).