On January 11, 2024, Plant Communications published a research paper titled "LbCas12a nuclease-mediated tiling deletion for large-scale targeted editing of non-coding regions in rice". This study proposed a LbCas12a nuclease-mediated tiling deletion (NTD) strategy, and used the rice plant height gene SD1 regulatory region as a target to demonstrate the concept of this strategy.
The spatiotemporal expression of genes is critical to growth, development and stress response, and is also precisely regulated by non-coding regulatory regions of the genome that may be located in promoters, introns and other regions. To date, a large number of natural or mutagenized variants in non-coding regions have been reported to contribute to crop domestication and breeding. The fine-tuning of gene expression achieved by editing non-coding regions may lead to quantitative trait variation and can screen a more balanced combination of agronomic traits from the quantitative trait variation population, so it is very suitable for crop improvement.
The technology currently used to edit non-coding regions of endogenous genes is mainly CRISPR/Cas9-mediated gene editing technology. However, compared to Cas9, LbCas12a tends to recognize the PAM motif of TTTV and induces larger deletions in plant cells. Due to the enrichment of A-T nucleotides and highly repetitive sequences in the non-coding regions of rice endogenous genes, LbCas12a-mediated NTD technology is more conducive to editing regulatory elements in non-coding regions and is more suitable for genetic engineering in non-coding regions.
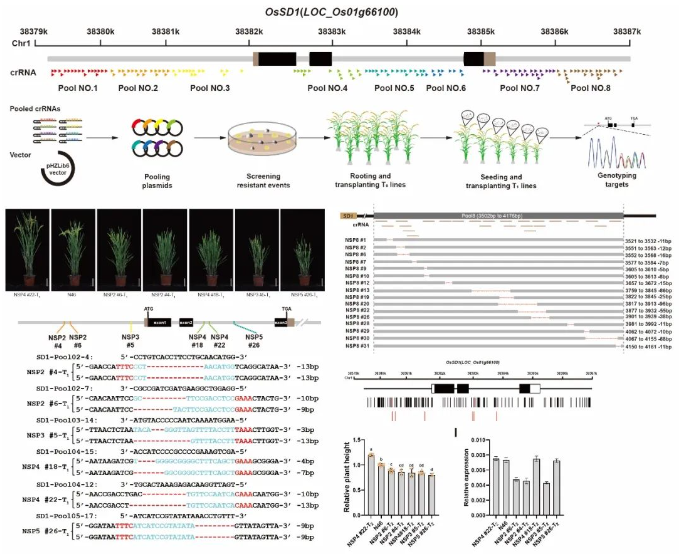
Based on the principle of NTD, the researchers carried out saturated coverage of the promoter, intron, terminator and other non-coding regions of the Green Revolution gene SD1, and designed and constructed a crRNA library for large-scale targeting. The non-coding region was targeted by Agrobacterium-mediated genetic transformation, and a total of 255 T0 mutants were screened and identified. Effective coverage editing can be achieved in different non-coding regions, and the mutation types are mainly deletions of 6-15 bp. Six materials with quantitative trait changes in plant height were also successfully screened and identified in the T1 generation. Further analysis of the representative phenotype, expression level, and T-DNA isolation of T2 determined that these quantitative trait variations were caused by editing of non-coding regions. Among them, 4 mutation sites were not detected in the resequencing data of 4726 natural populations of rice. In summary, it is shown that NTD technology can effectively create mutant populations with quantitative traits and screen and create regulatory elements for alleles in non-coding regions.
The proposal of NTD technology and its successful application in rice indicate that in the future, the expression levels of genes related to key crop traits can be fine-tuned and optimal agronomic trait combinations can be discovered. As a result, various agronomic traits of each crop, such as plant height, plant type, disease and insect resistance, etc., can be customized for expression control. NTD technology complements the plant endogenous gene directed evolution technology (base-editing-mediated gene evolution, BEMGE) previously reported by the team, and is expected to become a key tool for precision molecular breeding of crops in the future. In addition, the application of NTD technology can also promote the study of gene function regulation mechanisms in modern molecular biology, help people gain a deeper understanding of the mysteries of gene expression regulation, and can be further applied to molecular breeding.
Reference:
Ma, G., et al. LbCas12a nuclease-mediated tiling deletion for large-scale targeted editing of non-coding regions in rice. Plant Commun. 2024: 100815.