Lifeasible fuels the application of gene editing technology in plant protection and provides CRISPR/Cas9 editing services to edit plant genes. We can achieve efficient plant gene editing with CRISPR/Cas9 technology.
In 2012, CRISPR/Cas9 technology was discovered as a gene editing tool. Based on its ease of operation, high targeting ability, and high editing efficiency, CRISPR/Cas9 technology has developed a wide range of applications in biology, medicine, and agronomy. CRISPR/Cas9 has been successfully applied to various plants, including model plants (e.g., Arabidopsi, tobacco, and rice) and non-model plants (e.g., wheat, maize, and banana). CRISPR/Cas9 technology has high editing efficiency and can simultaneously edit multiple target genes. In addition, it can induce specific fragment deletions between target sites. The CRISPR/Cas9 system has two main components: the Cas9 protein and the single guide RNA (sgRNA). Designing sgRNA, selecting suitable Cas9 codons and promoters for specific plants, and selecting appropriate vectors and transformation methods are required to improve the efficiency of CRISPR/Cas9 technology in plant editing.
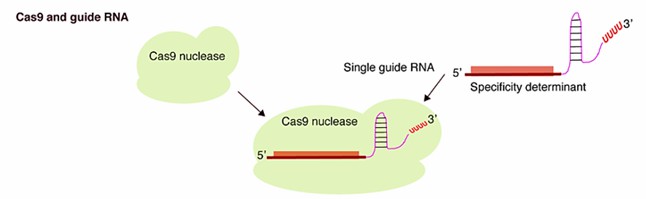 Fig. 1 The CRISPR/Cas9 system consists of a Cas9 protein and one or several guide RNA (El-Mounadi et al., 2020).
Fig. 1 The CRISPR/Cas9 system consists of a Cas9 protein and one or several guide RNA (El-Mounadi et al., 2020).
Lifeasible can provide plant gene editing using Streptococcus pyogenes Cas9 (SpCas9) and its related engineered variants, such as xCas9, SpCas9-NG, and SpCas9-VQR. Among them, we mainly use SpCas9 for gene editing. Below are the advantages of our gene-editing services with SpCas9.
Efficient sgRNA design
The secondary structure of sgRNA affects its binding and binding stability to Cas9 protein. Well-designed sgRNAs can improve the efficiency of plant editing. We provide standardized and efficient sgRNA design services to enhance plant gene editing efficiency. We help design sgRNAs to meet the following requirements as much as possible to ensure editing efficiency.
Codon-optimized Cas9
Enhancing the expression of Cas9 protein helps improve plant editing efficiency. We offer multiple codon-optimized Cas9 for plant gene editing and help select the appropriate codon-optimized Cas9 according to the plant species.
Plant-based selection of promoters
Strong promoters can be selected to boost sgRNA expression. We help to select strong promoters to boost sgRNA expression. For example, we offer to select promoters from rice (e.g., OsU6a, OsU6b, OsU6c, and OsU3) for monocotyledonous and dicotyledonous plants and promoters from Arabidopsis (e.g., AtU6-1, AtU6-29, AtU3b, and AtU3d) for Brassicaceae plants. Customers can also choose promoters according to their needs.
Efficient plasmid construction
We mainly use Golden Gate cloning for plasmid construction, which is a convenient and efficient way to clone complex plasmid vectors. This facilitates the expression of multiple sgRNA cassettes to edit multiple sites in the plant.
Efficient transformation
We mainly use Agrobacterium binary vector system for plant transformation. This system has high transformation efficiency and can obtain marker-free transgenic plants.
Obtaining non-Cas9-transformed gene-edited plants
We offer multiple ways to help get non-cas9-transformed gene-edited plants, including transient expression of CRISPR/Cas9, Mendelian segregation, programmed self-elimination of transgenic plants, and ribonucleoprotein-mediated genome editing.
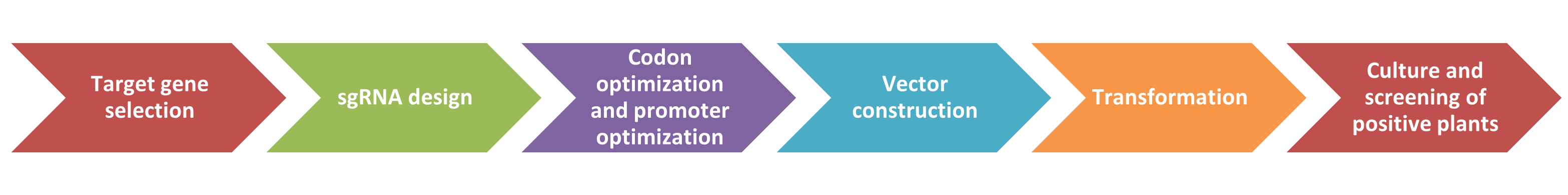
Lifeasible assists in applying CRISPR/Cas9 technology in plant gene editing. Please contact us for professional service.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024