Plant metabolome can be divided into primary metabolome and secondary metabolome. The primary metabolome is a collection of primary metabolites that are necessary for plant growth and development, while the secondary metabolome is a collection of secondary metabolites that often have special functions and can help plants adapt to special environments and resist pests and diseases. Not only that, many secondary metabolites are of great value to agriculture, medicine and industrial production, and provide abundant raw materials for human beings. Therefore, dissecting the biosynthetic pathways and genetic mechanisms of plant secondary metabolites is crucial for understanding basic biology, transformation and utilization of plant metabolic engineering.
Genome-wide association analysis (GWAS) is a method to locate genetic loci associated with traits by examining the significance of genome-wide genetic markers (such as SNPs) associated with phenotypic variation, and to analyze the genetic basis of traits at the population level. If the phenotype of interest is metabolite abundance, the process is called metabolite GWAS (mGWAS). mGWAS has become a valuable tool for the combination of metabolomics and genomics to identify new metabolic genes and discover new metabolic pathways. It has been widely used in the model plant Arabidopsis thaliana and many crops (including rice, corn, wheat and tomato). Nevertheless, conventional mGWAS still has two limitations: first, when performing GWAS calculations, the genotype frequency of the selected genetic markers is greater than 5%, so the variants with a gene frequency less than 5% are not included, resulting in some rare variants being ignored; second, GWAS results are affected by the population structure, and an inappropriate population structure can easily cause false positives.
On June 21, 2023, Wout Boerjan's research group at Ghent University and VIB Plant Systems Biology (PSB) published a research paper entitled "QT-GWAS: a novel method for unveiling biosynthetic loci affecting qualitative metabolic traits" in Molecular Plant, proposing a new genome-wide association analysis method (QT-GWAS) based on qualitative metabolic traits. QT-GWAS not only includes the variation of gene frequency less than 5%, but also does not need to consider the population structure. Compared with the traditional mGWAS method, QT-GWAS has a stronger localization effect and more loci.
Qualitative and Quantitative Traits
Metabolite qualitative traits refer to metabolites that are absent in some individuals but present in others in a particular population, and the presence/absence of such metabolites can be considered a qualitative rather than quantitative trait. In this study, 183 Arabidopsis thaliana of different ecotypes were firstly measured by non-targeted metabolome, and a total of 5082 metabolic features were detected. Among them, 4479 features were defined as qualitative traits, 1147 features were defined as quantitative traits, and 702 features belonged to both qualitative and quantitative traits.
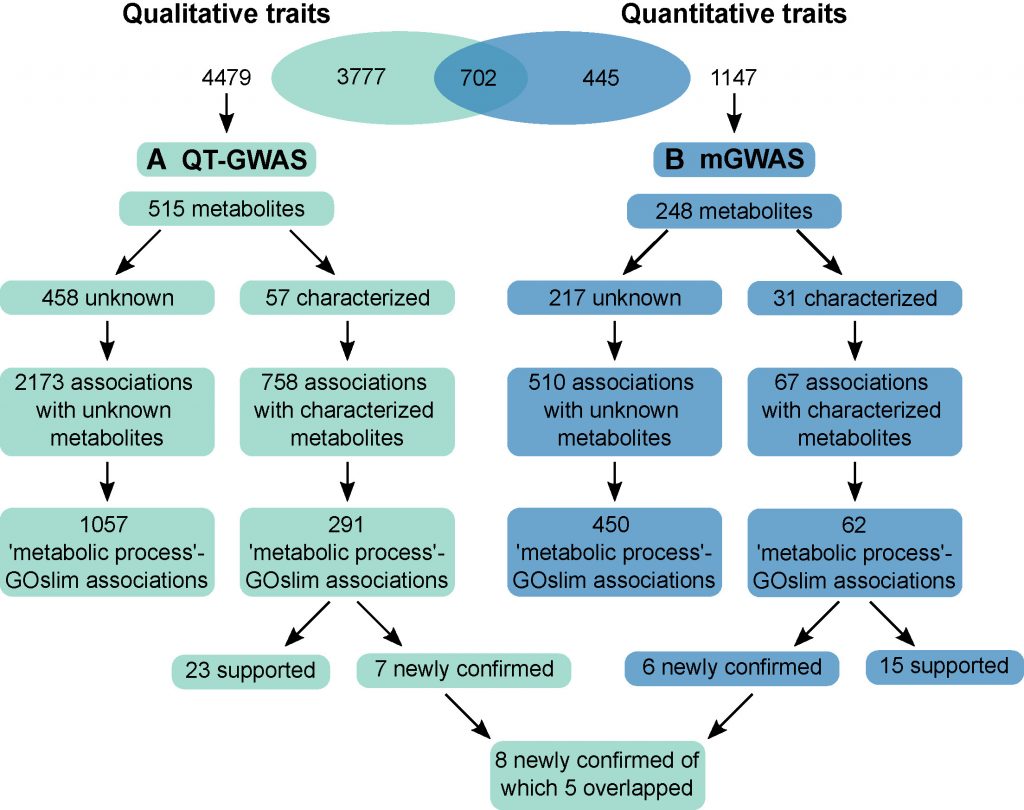
Comparison of QT-GWAS and mGWAS Calculation Process and Positioning Results
The association analysis between quantitative traits and SNPs used Fisher’s exact test to calculate the significance between 4479 quantitative features and each SNP, and set the threshold as 10-6. After screening, 515 metabolites obtained 2931 significant associations. Of the 515 metabolites, 397 (77%) were associated with one locus and 118 (23%) were associated with multiple loci. A total of 23 pleiotropic loci were found out of 2931 loci.
The association analysis between quantitative metabolic traits and SNPs adopts the conventional GWAS analysis process. After screening, a total of 577 significant associations were obtained from 248 metabolites. Of the 248 metabolites, 223 (90%) were associated with one locus, while 25 (10%) were associated with multiple loci. A total of 6 pleiotropic loci were found out of 577 loci, 2 of which coincided with QT-GWAS.
In order to investigate whether loci associated with metabolic traits are enriched in genes involved in metabolic processes, the researchers performed a GOterm enrichment analysis on the results mapped by these two methods using PANTHER. For QT-GWAS, 103 GOterms were significantly enriched in the category of “Biological Processes”, of which 22 GOterms were related to various metabolic processes. For conventional mGWAS, 25 GOterms were significantly enriched, 5 of which were involved in metabolic processes. Notably, these 5 metabolic process-related GOterms were more significantly enriched in QT-GWAS, although the fold-change sizes of the enrichments were similar. In order to further compare the positioning results of QT-GWAS and mGWAS, the authors compared the co-localization results of 702 traits in QT-GWAS and mGWAS. Among them, 71 traits were associated in both QT-GWAS and mGWAS, and the overlap coefficient of the association results was only 0.32, which means that only 32% of the genes retrieved by mGWAS overlapped with those retrieved by QT-GWAS, indicating that QT-GWAS can retrieve new associations not found by mGWAS.
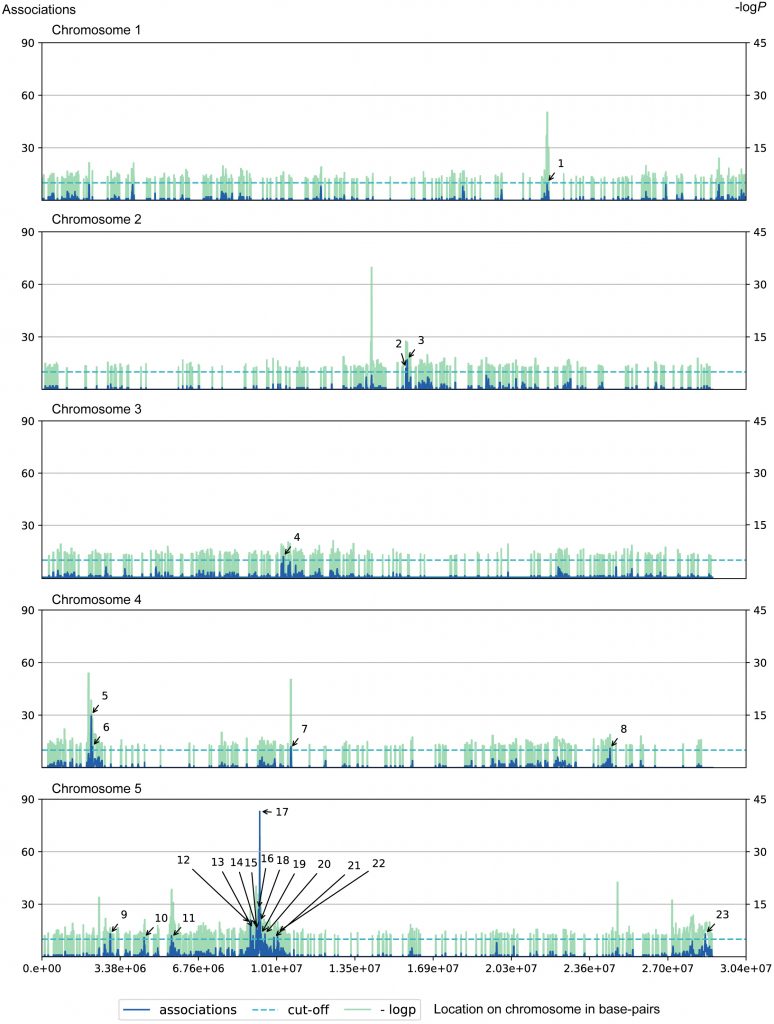
Functional Verification of Candidate Genes
Using QT-GWAS, the researchers not only located 26 reported loci, but also located 3 new loci, and used reverse genetics to prove the functions of 3 candidate genes: CYP706A5, a gene involved in chroman biosynthesis; UGT67C3, a gene involved in guanine glycosylation; SULT202B1, a gene involved in neolignan sulfation.
In conclusion, this study established a genome-wide association analysis method based on quantitative traits of metabolites (QT-GWAS), which overcomes the limitations of conventional mGWAS, further improves the effect of metabolite mapping, and may locate more candidate genes. If QT-GWAS is reapplied to previously published mGWAS data, it is highly possible to discover new results. If the two methods are combined, it should be possible to further enhance the potential of GWAS methods to mine new genes and new pathways of secondary metabolism within the system.
Reference:
Brouckaert, M., Peng, M., Höfer, R. et al. QT-GWAS: A novel method for unveiling biosynthetic loci affecting qualitative metabolic traits. Mol Plant. 2023, 16(7): 1212-1227.