On June 6, 2025, Seiichi Toki's team from the Japan Agricultural and Food Research Institute published a new research paper titled "Heritable Large Deletions Using Type I-E CRISPR-Cas3 in Rice" on bioRxiv. This study successfully achieved heritable large gene deletion editing mediated by Type I-E CRISPR-Cas3 in rice for the first time. This breakthrough has added an important new member to the plant gene editing toolbox.
Compared with the familiar CRISPR-Cas9 system, the application of Type I CRISPR-Cas system in plants has always faced great challenges. The research team pointed out that the Type I-E CRISPR-Cas3 system requires the simultaneous expression of 7 components (6 Cas proteins and CRISPR RNA), which makes its application in plant cells extremely difficult.
Previously, the application of Cas3 in plants was limited to corn protoplasts, and mutant plants had never been obtained. This has become an important bottleneck restricting the promotion and application of this technology.
To overcome these difficulties, the research team developed a clever combination of strategies.
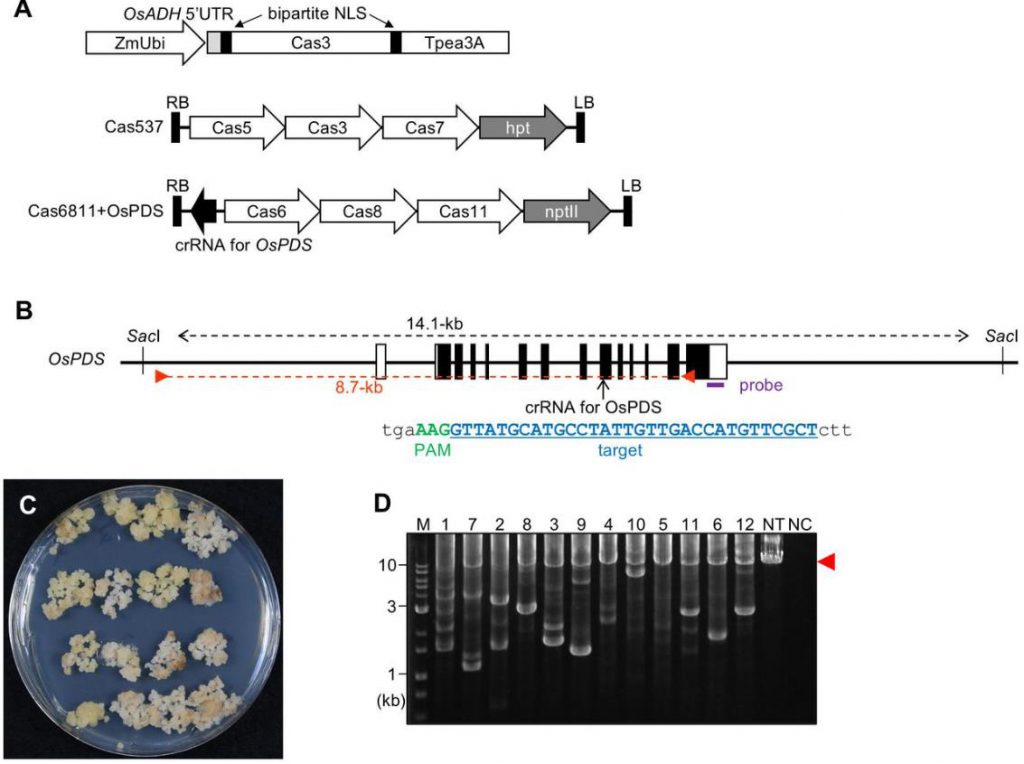
Figure 1. Eco CRISPR-Cas3-mediated targeted mutagenesis in rice calli. (Saika, et al., 2025)
The study revealed significant differences between Cas3 and traditional Cas9 systems.
Sequence analysis showed that Cas3 can not only produce simple large deletions, but also create new alleles such as complex deletions separated by intervening fragments, deletions accompanied by short insertions and inversions.
The research team verified the editing efficiency of Cas3 by multiple methods.
Using droplet digital PCR (ddPCR) technology, the researchers accurately determined the deletion frequency at different positions:
The most important breakthrough of the study is that it proves that large deletions mediated by Cas3 can be stably inherited by offspring.
In the OsWx gene editing experiment, the researchers obtained a T0 plant with a 2.8 kb deletion, all 12 of whose T1 seeds showed a waxy phenotype, and PCR analysis confirmed the stable inheritance of biallelic mutations.
This result shows that Cas3 can produce biallelic mutant plants with an efficiency of 14%, providing a new and powerful tool for plant breeding.
The research team pointed out that the unique large deletion characteristics of Cas3 make it a promising tool for gene knockout, gene deletion and genome rearrangement. Compared with the traditional method that requires the design of dozens of gRNAs to achieve large promoter deletions, Cas3 only needs a single gRNA to create diverse promoter alleles.
This study established a complete I-E type CRISPR-Cas3 gene editing system in plants for the first time, which not only expanded the plant gene editing toolbox, but also opened up a new technical path for crop precision improvement and molecular breeding. With the further optimization and promotion of this technology, we have reason to expect more breakthrough applications.
| Cat# | Product Name | Size |
| ACC-100 | GV3101 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-103 | EHA105 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-105 | AGL1 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-107 | LBA4404 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-108 | EHA101 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-117 | Ar.Qual Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-118 | MSU440 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-119 | C58C1 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-121 | K599 Chemically Competent Cell | 10 tubes (100μL/tube) 20 tubes (100μL/tube) 50 tubes (100μL/tube) 100 tubes (100μL/tube) |
| ACC-122 | Ar.A4 Electroporation Competent Cell | 10 tubes (50μL/tube) 20 tubes (50μL/tube) 50 tubes (50μL/tube) |