Grasslands cover a large part of the world's agricultural area and play an essential role in forage production for herbivores, as well as being providers of ecological services to protect water quality, biodiversity, and carbon sequestration. Some grasslands are mainly used for ruminant forage production and are regularly sown with pasture and legumes. Most forage species are exclusively distantly bred. Their breeding usually involves the initial selection of plants grown at intervals for highly genetic traits such as disease resistance and tassel stage, followed by a familial selection of grasses for forage yield and quality traits. Most traits in forage grasses are complex and regulated at the genomic, transcriptomic, epigenetic, proteomic, gene interaction, and regulatory network levels. However, in practical forage breeding programs, only genealogical and genomic data are currently routinely used, which often need more explanations of genetic variation. The concept of genomic prediction was derived as an alternative to marker-assisted selection.
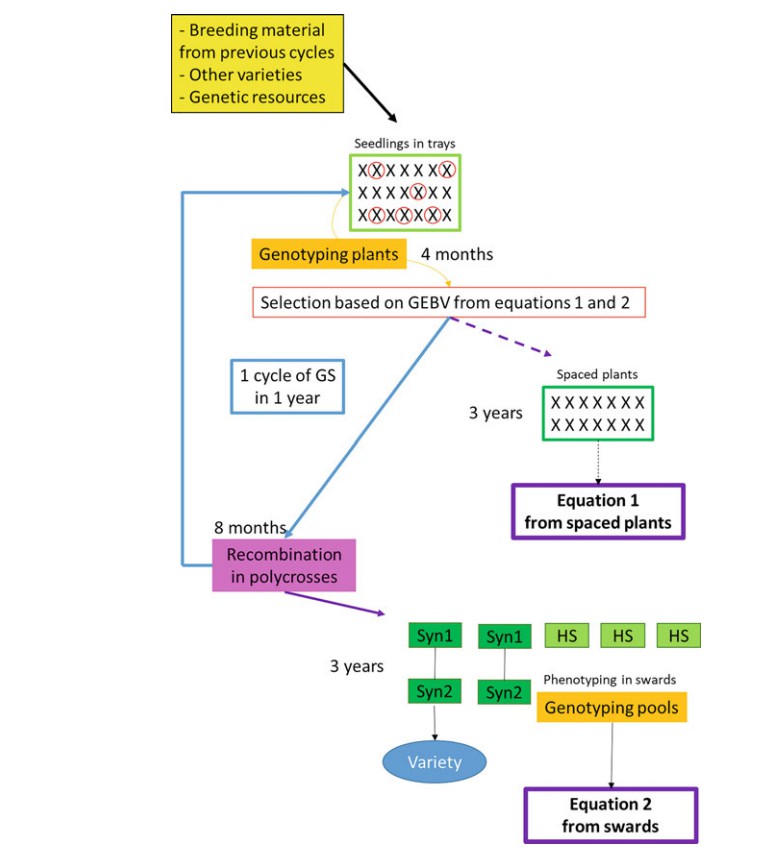 Fig.1. A breeding scheme for forage grass species integrating genomic selection. (Barre P et al., 2022)
Fig.1. A breeding scheme for forage grass species integrating genomic selection. (Barre P et al., 2022)
To accurately predict complex traits in forage breeding, our scientists work to identify and explain the origin of the variation behind these traits. As the ideal partner for forage research, Lifeasible provides a comprehensive genomic prediction service for forage dominant species to clients worldwide, allowing the generation of molecular and phenotypic information in a high-throughput manner and the integration of all this data with information on management practices using machine learning and artificial intelligence. Our deep learning approach facilitates the development of multi-omics data strategies to improve predictive models in forage breeding.
We provide a traditional genomic prediction pipeline involving the development of a training set of available genotypic and phenotypic data for constructing a prediction model. In addition, the model is used to estimate breeding values from the genomes of individuals selected as candidates in the prediction validation set, which consists of genotyped but unphenotyped individuals. We calibrated the model by implementing a cross-validation scheme, taking subsamples from the training set, and then fitting the model to the rest of the training set to estimate and evaluate its predictive power.
We provide genome-wide family predictions to screen traits for phenotypic typing in forage grasses. Here, allele frequencies and phenotypic records are expressed as a single average record for a given family. Thus, the additive genetic variance in a full-sibling family is half the additive variance between individuals. Predictive power was higher in the family pool compared to the genomic estimated breeding values for individuals.
Forage grass is a polyploid species with higher genomic complexity and limited genetic resources, hindering the application of marker-assisted selection strategies. We develop a machine learning approach to predict complex traits from genotype data using a joint learning approach. In addition, we provide high-quality markers to predict several traits in different cross-validation scenarios.
Our advanced next-generation sequencing technologies combined with genomic resources improve forage dominant species genomic prediction accuracy by providing forage-associated intermediate-level information to elucidate the association between genotype and phenotype. Our breeders use a sufficient number of markers to cover the entire forage genome, and prediction models are trained in appropriate environments. Lifeasible has the theory and tools for gene selection to be replicated in forage species breeding programs. For more information or to discuss in detail, please contact us.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Why Do Plants Blush When They Are Hungry?
April 26, 2024
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024