Cotton, including Upland cotton (Gossypium hirsutum) and Sea Island cotton (Gossypium barbadense), is a commercial crop of world importance. It is the world's major textile fiber crop and a source of minor products such as oil, live animal feed (cottonseed cake), and cellulose. Although classical breeding has contributed greatly to quality improvements and increased yields in the twentieth century, textiles require further improvements in fiber strength, length, chemical compatibility of dye binding, water absorption, thermal properties, and resistance to wrinkles and shrinkage sex. The potential to improve these traits through classical breeding is limited due to requirements for species compatibility and availability of traits. Genetic engineering will provide new methods to overcome the shortcomings of classical breeding by introducing target genes from different sources into cotton. However, there are fewer studies on the integration structure of exogenous genes in transgenic cotton, the molecular characterization of exogenous insertion fragments, etc. Figuring out the integration structure of exogenous genes in transgenic cotton will help to establish a detection method for transgenic cotton, which is of great significance for transgenic biosafety.
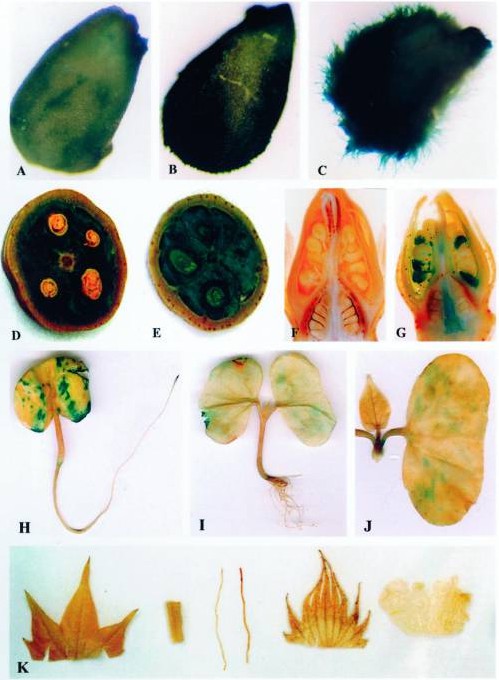 Fig. 1. Histochemical localization of GUS gene expression in transgenic cotton plants containing the GhTUB1/GUS fusion gene. (Li et al., 2002)
Fig. 1. Histochemical localization of GUS gene expression in transgenic cotton plants containing the GhTUB1/GUS fusion gene. (Li et al., 2002)

We provide advanced plant genetic engineering services to improve cotton genetic traits by applying gene gun, pollen pipeline, Agrobacterium-mediated methods, etc. We can help you breed high-quality insect-resistant, herbicide-resistant cotton, etc. With years of experience, Lifeasible provides professional cotton molecular characterization analysis services to evaluate the safety of genetically modified cotton. We can provide you with cotton molecular characteristics information at the chromosome level, including the T-DNA insertion site, copy number, and flanking sequences of foreign genes.
We have a complete genetically modified corn molecular characterization detection system, including Southern hybridization, Polymerase chain reaction (PCR) analysis, PCR-based genome walking, quantitative real-time polymerase chain reaction (qRT-PCR), Southern hybridization, Western blotting, ELISA, high throughput sequencing, etc. Based on your project requirements, we will select the most suitable detection method for analyzing molecular characterization of genetically modified cotton. We can analyze the following genetically modified cotton, including but not limited to:
| GhTUB1 genetically modified cotton | GhTUA9 genetically modified cotton | 14-3-3L genetically modified cotton | Deltapine Acala 90 phytase genetically modified cotton |
| Deltapine 41 genetically modified cotton | DES 422 genetically modified cotton | Protease inhibitor genetically modified cotton | GhTUA9 genetically modified cotton |
| GV-2 genetically modified cotton | GhMKK9 genetically modified cotton | R1-3 genetically modified cotton | Bt-Cry5Aa genetically modified cotton |
| GNA genetically modified cotton | CPTI genetically modified cotton | Cry 1Ac genetically modified cotton | Cry1Ab genetically modified cotton |
| Cry1F genetically modified cotton | Cry2Ab genetically modified cotton | Cry2Ae genetically modified cotton | Vip3A(a) genetically modified cotton |
| S4-HrA genetically modified cotton | Epsps genetically modified cotton | Pat genetically modified cotton | Bar genetically modified cotton |
| Bxn genetically modified cotton | Dicamba O-demethylase genetically modified cotton | - | - |
Work Process of Genetically Modified Cotton Molecular Characterization Analysis
Lifeasible aims to obtain the integrated structure and molecular characteristics of exogenous genes in genetically modified cotton and to establish the specific detection method for transformation events and the screening method for pure congeners. Please contact us for the best analysis solutions for genetically modified cotton molecular characterization.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024