Insects are the most diverse and numerous of the animal kingdom. They are found in almost every corner of the world and significantly impact agricultural production and human health. Insects are easy to breed, have a short life cycle, and can obtain a large number of individuals in a short period of time; they are open-cycle animals, and transplantation of organs and endocrine glands is relatively easy, and many physiological problems of invertebrates are studied using insects as experimental materials.
With the development of sequencing technologies and the reduction of sequencing prices, more and more insect genomes are being decoded, which provides the basis for developing insect epigenetic sequencing technologies, such as CHIP-seq. As scientists continue to expand the scope of insect research, the application of CHIP-seq in insects is becoming more and more common, providing new ideas and methods for studying the entire range of biological laws, such as development, ecology, and genetics.
Lifeasible has a mature platform and extensive experience in insect sequencing and can provide customers with insect ChIP-Seq solutions for a wide range of species, such as silkworms, drosophila, and mosquito.

We use antibodies specific to histone-specific modifications or DNA binding proteins, or transcription factor-specific antibodies to enrich for DNA fragments bound to them, then build libraries and sequence them. This service combines second-generation sequencing to search for genome-wide binding sites for transcription factors, epitope-modifying enzymes, or DNA-binding proteins.
To decipher the regulatory role of the transcription factor Broad Complex (BR-C) in insect growth and development, we performed whole-genome sequencing of BR-C target genes in silkworm (Bombyx mori) using ChIP-seq. The results showed that BR-C was enriched in pathways related to biosynthesis, metabolism, and development.
To characterize lncRNAs in Drosophila melanogaster, we developed a novel protocol to detect active transcription of lncRNAs using ChIP-seq data (H3K4me3, H3K36me3, and Pol II). This study provides a basis for exploring the function of lncRNAs in Drosophila melanogaster.
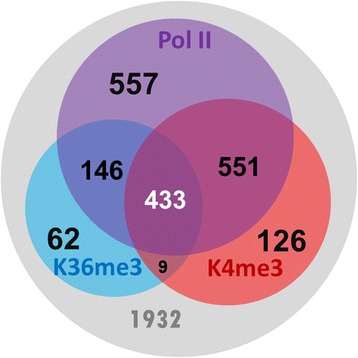 Figure 1. Analysis of chromatin signatures (Pol II, H3K36me3, and H3K4me3) in the curated lncRNA genes. (Chen, M. J, et al. 2016)
Figure 1. Analysis of chromatin signatures (Pol II, H3K36me3, and H3K4me3) in the curated lncRNA genes. (Chen, M. J, et al. 2016)
To investigate the potential mechanisms underlying the changes in vector competence induced by malaria infection in mosquitoes, we used ChIP-seq to sequence H3K27ac, H3K9ac, H3K9me3, and H3K4me3 at the whole genome level. The results of the data analysis helped us to analyze the regulatory elements and genes of mosquitoes in response to P. falciparum infection.
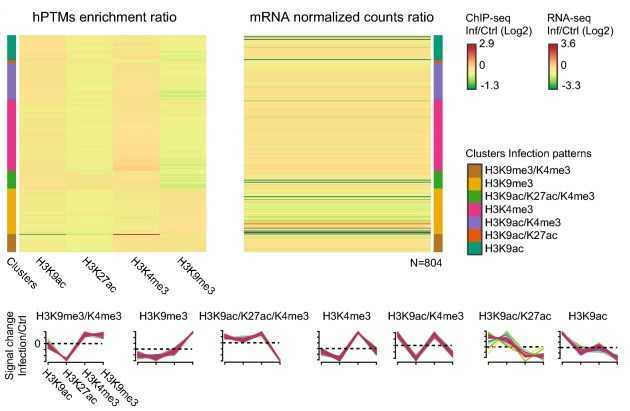 Figure 2. Analysis of histone ChIP-seq data. (Ruiz, J. L, et al. 2019)
Figure 2. Analysis of histone ChIP-seq data. (Ruiz, J. L, et al. 2019)
It is recommended to provide two sample preparations, if possible, to ensure the quality and continuity of the experiment.
Lifeasible's animal-oriented sequencing technology platform can provide complete insect ChIP-seq services to our customers. We are committed to meeting all of our customer's needs, and we guarantee to obtain high-quality reads and perform comprehensive bioinformatic analysis. Please feel free to contact us for any questions, inquiries, or cooperation.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024