Almost all human genes associated with the disease can find counterparts in the rat genome. In contrast, rats are several times larger than mice and possess fewer stress responses, enabling better research on disease mechanisms, new drug evaluation, etc.
As epigenomic sequencing technology continues to be applied in rats, more epigenetic regulatory mechanisms are being elucidated. ATAC-seq technology can detect chromatin openings genome-wide in rats and obtain genome-wide information on possible protein binding sites, helping researchers to identify valuable transcriptional regulatory regions.
Lifeasible focuses on providing efficient and accurate sequencing services. Our professional high-throughput sequencing platform can provide quality basic data for disease mechanism research and drug discovery. We offer a wide range of rat ATAC-seq technology solutions that can effectively advance our clients' projects and help them explore the state of binding of various regulatory factors to open chromatin.

We sequenced rat skeletal muscle-associated genes using single nucleus ATAC-seq (snATAC-seq) technology to map specific chromatin accessibility. Skeletal muscle cell types were identified by multi-omics analysis with sequencing results from human cells. Moreover, we identified SNPs, transcription factors, and target genes associated with diabetic signaling and further went through cell types to discover the relationship between cells and diseases.
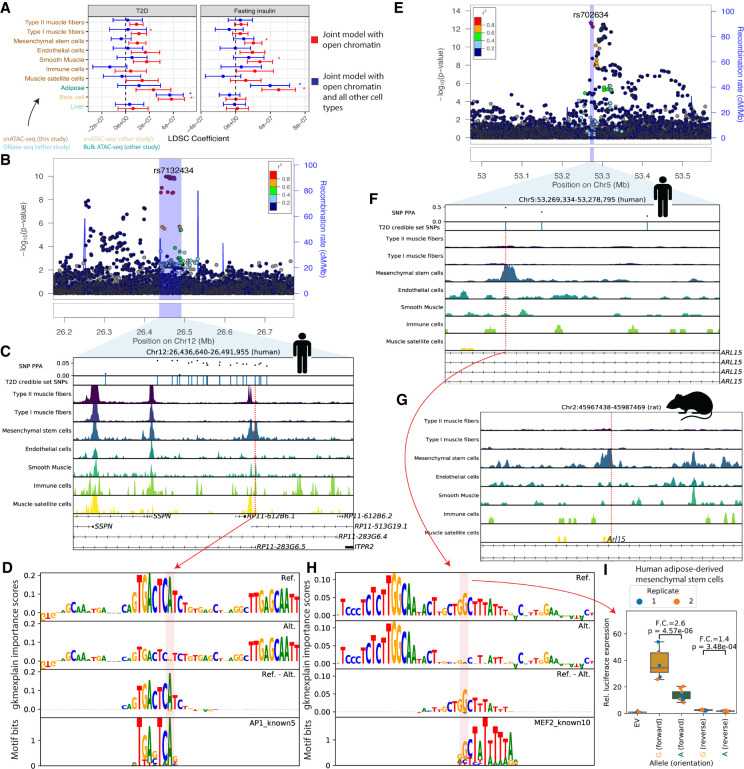 Figure 1. Cell type-specific ATAC-seq peaks and the cell types, regulatory elements, and SNPs were analyzed. (Orchard, P et al. 2021)
Figure 1. Cell type-specific ATAC-seq peaks and the cell types, regulatory elements, and SNPs were analyzed. (Orchard, P et al. 2021)
We have optimized the reproducibility of ATAC-seq analysis in adipocytes to enable rapid and accurate chromatin genomic analysis and transcription factor binding site discovery in rat adipose tissue. Our experimental data are highly reproducible, helping scientists to advance relevant exploratory research in adipose tissue.
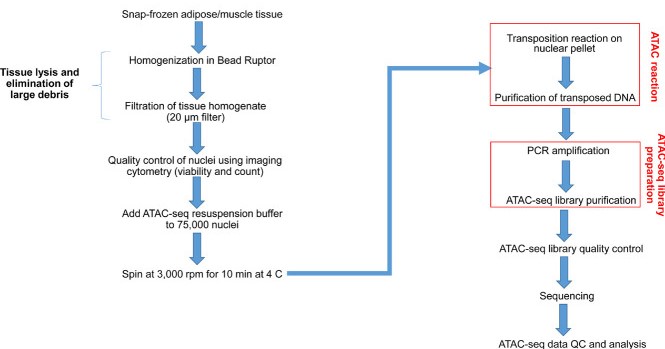 Figure 2. Flow chart of chromatin accessibility analysis of rat adipose and muscle tissue using ATAC-seq. (Nair, V. D, et al. 2022)
Figure 2. Flow chart of chromatin accessibility analysis of rat adipose and muscle tissue using ATAC-seq. (Nair, V. D, et al. 2022)
We used ATAC-seq technology to examine the transcriptional profile of genes involved in the peripheral nervous system in a rat model of chronic pain response and the epigenetic basis of chronic pain. Different pain models demonstrate changes in chromatin accessibility, providing a good idea for identifying pathways associated with pain response and facilitating scientists to identify regulatory elements that play an essential role in persistent pain.
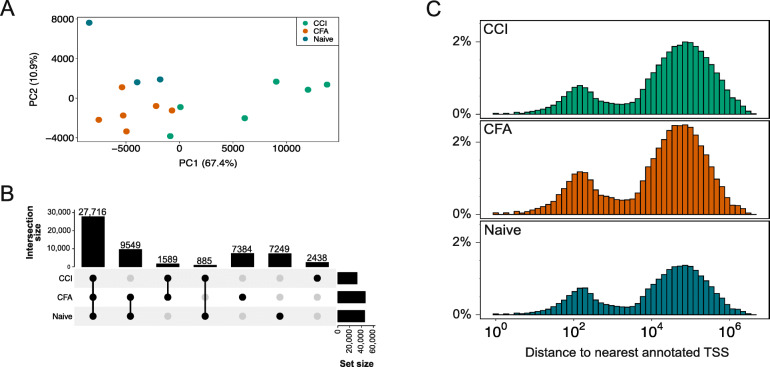 Figure 3. Chromatin accessibility in rat DRG. (Stephens, K. E, et al. 2021)
Figure 3. Chromatin accessibility in rat DRG. (Stephens, K. E, et al. 2021)
Technical Route of Rat ATAC-Seq
Please note: Ensure sufficient dry ice when transporting samples to avoid temperature rise to destroy cell activity.
Lifeasible provides professional rat ATAC-seq services according to our client's needs. Our state-of-the-art laboratory facilities, well-established animal houses, and rigorous sequencing data management can ensure data stability between multiple batches, providing our clients with authentic and accurate data. We focus on saving our clients' time and effort, allowing them to focus on their most scientifically innovative work. Please feel free to contact us with questions, inquiries, or collaborations.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024