The use of organelle genomic information has become a common practice in comparative genome-wide studies and phylogenetic analyses. In eukaryotic lineages, mitochondrial genomes are highly variable in size, structure, and gene content. Studies of targeted sequencing of mitochondrial genomes from under-sampled lineages have allowed a better understanding of the evolutionary history of mitochondrial genomes in specific taxa. Most microalgae contain four distinct genomes: the nuclear genome, the nuclear morphogenome, the mitochondrial genome, and the plastid genome. Therefore, microalgae are considered an interesting model for studying genome evolution and protein targeting. The mitochondrial genome of Chlamydomonas reinhardtii (unicellular green algae) is a linear molecule of 15.8 kbp. In terms of gene arrangement, expression patterns, and size, it is quite different from the large (200-2400 kbp) mitochondrial genomes of higher plants.
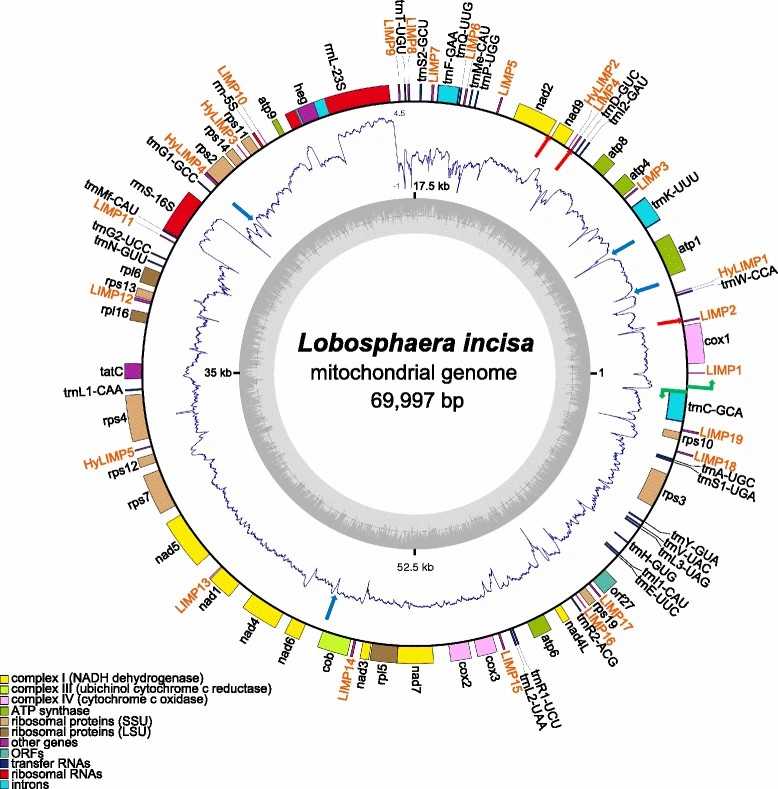 Fig 1. Schematic representation of the L. incisa mitochondrial genome. (Tourasse N J, et al., 2015)
Fig 1. Schematic representation of the L. incisa mitochondrial genome. (Tourasse N J, et al., 2015)
Green plants' mitochondrial genome (mtDNA) is of interest because of its complex genome and structural diversity across evolutionary branches. Sequencing of mitochondrial genomes can provide valuable information on the evolution of organelles and the organisms that carry them. Based on our extensive experience in microalgal genomics, Lifeasible provides professional microalgae mitochondrial genomics services for clients to sequence, de novo assembled, and functionally annotate the complete mitochondrial genomes of a wide range of microalgae. We have developed customized mitochondrial genomics processes for microalgae.
(1) Algal biology isolation and culture.
(2) DNA extraction and purification according to standard protocols.
(3) Library construction and sequencing and assembly of the mitochondrial genome.
(4) Mitochondrial genome annotation.
(5) Phylogenetic analysis of the mitochondrial genome.
(6) Perform genome-wide comparisons.
Mitochondrial genomes throughout eukaryotes vary significantly in size, gene content, and genome organization We aim to obtain complete microalgal mitochondrial genomes using high-throughput sequencing (HTS) technology and perform comparative analyses of genome structure and gene rearrangements.
Our solutions provide essential technical support for the broad evolution of organelle genomes and the fine dynamics within microalgae. We can quickly help you obtain the complete mitochondrial genomes of multiple microalgae for sequencing to explore new genomes to determine general trends in the evolution of microalgal mitochondrial genomes. If you are interested in our solutions, please contact us directly.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024