As a livestock species that entered human society earlier, equines play a significant role in agriculture, transportation, and recreation. Due to the different natural environmental conditions of their origin, equines possess several breeds that can meet the multiple needs of human society. Also, the different phenotypes imply a complex genome composition and are model organisms for scientists conducting species evolution studies, gene function studies, and genetics studies.
Modern mechanization is accelerating, and equines no longer significantly impact society, but they remain key organisms in anatomy, genealogy, and sports science. With the rapid development of high-throughput sequencing and bioinformatics research, scientists have extensively used sequencing to analyze the whole genome sequence of equines, resolve the molecular mechanisms that shape complex traits, and probe the mysteries of biological epigenetics.
Lifeasible has been deeply involved in genome sequencing research for many years and is committed to providing quality one-stop equine ChIP-seq services to researchers. From DNA purification to deep sequencing to bioinformatics analysis, our customizable services are designed to solve your epigenetic research challenges.

Our equine ChIP-seq service can effectively address the impact of tissue metabolites on experimental results, providing you with high-quality and high-throughput experimental data.
We used ChIP-Seq technology to functionally annotate genes involved in specific regulation in the equine genome, including histones including H3K4me1, H3K4me3, H3K27ac, and H3K27me3, by which we can further analyze the network of protein interactions, effectively helping equine scientists Identify the molecular mechanisms that affect equine reproduction, health, and trait characterization.
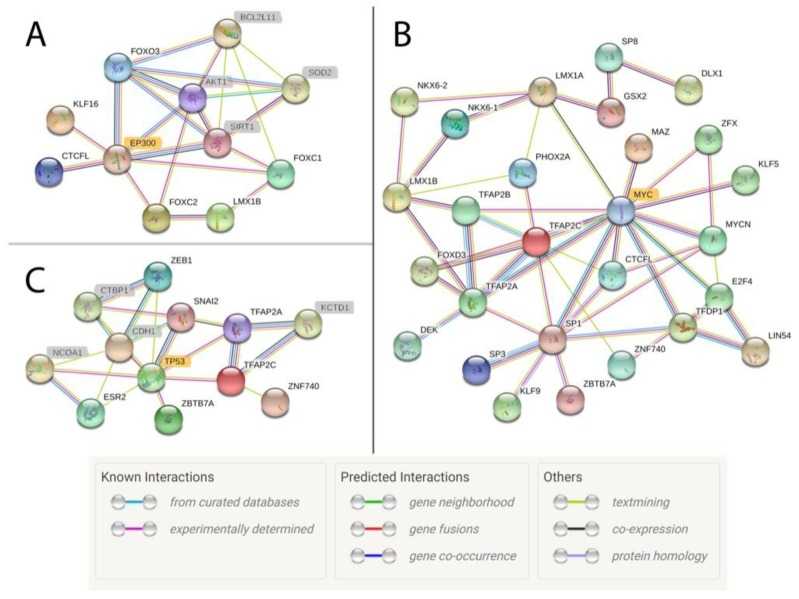 Figure 1. Transcription factor interaction network of active motif regions in ovarian, skeletal muscle, and lung tissues analyzed by CHIP-seq assay. (Kingsley, N. B, et al. 2019)
Figure 1. Transcription factor interaction network of active motif regions in ovarian, skeletal muscle, and lung tissues analyzed by CHIP-seq assay. (Kingsley, N. B, et al. 2019)
The absence of tandem repetitive sequences on the mitoses of equine chromosome 11 has been previously demonstrated. However, a common feature of these chromosomes is the presence of a satellite DNA family. Using ChIP-Seq technology, we found that the equine satellite DNA sequence 37cen binds specifically to histone CENP-A and demonstrated that the 37cen sequence is transcriptionally active.
It is recommended to provide two sample preparations, if possible, to ensure the quality and continuity of the experiment.
Lifeasible has accumulated a wealth of experience in animal sequencing services. We provide the most advanced equine ChIP-seq technology services to help you quickly reveal the secrets of equine histone modifications, DNA modifications, and chromatin conformation. We will be happy to serve you; please feel free to contact us if you need any help.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024