Bovines are one of the most critical agricultural animals and are extremely close to human life, being the main source of meat, milk, or hides. The genome sequence of bovines took many years to be deciphered. The bovine genome contains at least 22,000 genes, of which about 14,345 have counterparts in seven other mammalian species. The decipherment of the bovine genome will not only help us to gain a deeper understanding of the evolutionary and domestication processes of bovine. Still, it will also facilitate the application of epigenomics techniques, such as ChIP-seq, in bovine.
ChIP-seq technology is based on the Illumina Hiseq sequencing platform to study histone modifications or DNA regions bound by transcription factors on a genome-wide scale and to obtain high-throughput data results by efficient sequencing means. ChIP-seq technology can identify regulatory elements of the bovine genome and classify their functional components. The epigenome mapping by ChIP-seq can also help customers to study the biological systems, transcriptional regulation, and disease mechanisms of bovine.
Lifeasible's professional analytical team, rich project experience, and high-quality project services can provide excellent bovine ChIP-seq solutions for our clients to assist their scientific research fully.

Our bovine ChIP-seq service uses antibodies specific to histone-specific modifications or DNA binding proteins, or transcription factor-specific antibodies to enrich DNA fragments bound to them, purify and build libraries, and then perform high throughput sequencing.
Genetic variants that affect complex traits in bovines can link gene lists and phenotypes and are the focus of research by geneticists. We used ChIP-seq technology to examine the heart, kidney, liver, lung, mammary gland, and spleen of bovine. Ultimately, millions of functional regions in the bovine genome were determined, and genetic variants were found to be enriched in functional regions, demonstrating that genetic variants regulate complex traits.
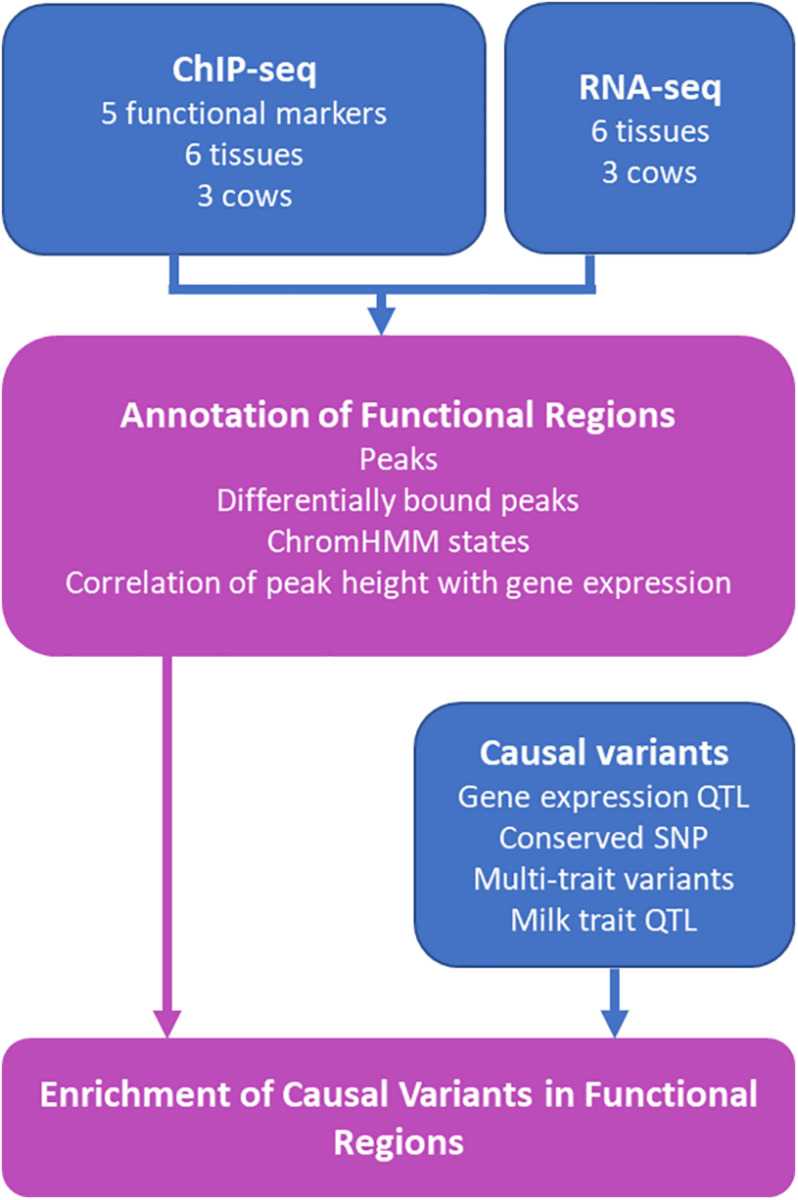 Figure 1. Schematic diagram of the study using ChIP-seq to detect putative causal variants in bovine tissues. (Prowse-Wilkins, C. P, et al. 2021)
Figure 1. Schematic diagram of the study using ChIP-seq to detect putative causal variants in bovine tissues. (Prowse-Wilkins, C. P, et al. 2021)
We examined the genome-wide binding motifs of 10 histone H3 and H4 modified variants using ChIP-seq. We analyzed ChIP-seq data to map the epigenomic landscape of these butyrate-induced histones in high resolution, thus revealing that butyrate-induced acetylation of H3K9 and H3K27 can alter the binding site preference of histone H3. Meanwhile, it provides a new way to analyze the epigenome's role in cellular function and transcriptome regulation.
ChIP-seq is an efficient method to detect DNA sites interacting with functional proteins at the gene-wide level. We used ChIP-seq to detect global mapping of the binding sites of phiC31 integrase in bovine kidney cells.
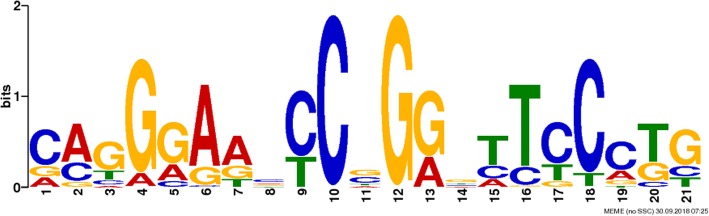 Figure 2. The phiC31 integrase-binding motifs were analyzed using ChIP-seq. (Qu, L, et al. 2019)
Figure 2. The phiC31 integrase-binding motifs were analyzed using ChIP-seq. (Qu, L, et al. 2019)
It is recommended to provide two sample preparations, if possible, to ensure the quality and continuity of the experiment.
Lifeasible's animal-oriented sequencing technology platform can provide efficient bovine ChIP-seq services to help customers obtain a high-definition map of the bovine epigenome, provide a basis for exploring the role of target genes in cellular function and transcriptome regulation, and deepen research on bovine evolution, reproduction, disease, morphology, and other fields. Please feel free to contact us for questions, inquiries, or collaboration.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024