The ocean is the birthplace of life, and the work of marine biology is indispensable to studying the origin and evolution of life. Marine invertebrates account for most marine animals and are a popular object of research in marine biology. Through research, scientists clarify the nature of life, the characteristics and habits of marine organisms, various biological phenomena occurring in the ocean, and their changing laws, and then use these laws to serve human life and production.
In recent years, the development of high-throughput sequencing technologies and the improvement of extensive data analysis capabilities have led to an increasing number of studies at the genomic level. More and more scientists are applying modern epigenetic techniques to study the epigenetic regulatory mechanisms of marine invertebrates adapting to extreme marine environments.
Lifeasible, as a specialized epigenome sequencing biology company, can provide high-throughput, customizable marine invertebrate ATAC-seq solutions to our clients. We use ATAC-seq technology to analyze genome-wide chromatin accessibility and identify signaling pathways and developmental cis-regulatory elements (CREs) for our clients.

To investigate the mechanisms regulating developmental processes in echinoderms, we optimized our experimental protocol and examined the chromatin of developing sea urchins using ATAC-seq. Combined with bioanalytical techniques, static assessment of chromatin structure revealed local, hyper-accessible regions and identified active cis-regulatory elements in sea urchins.
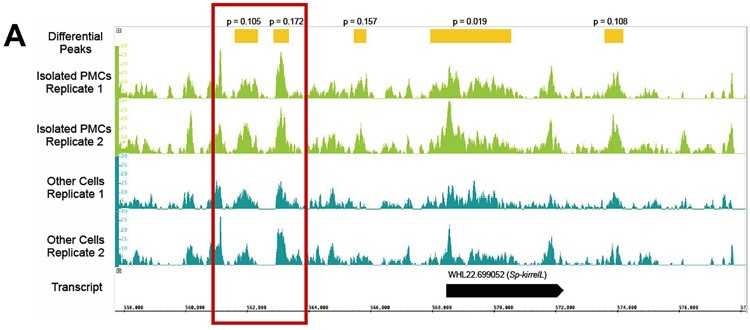 Figure 1. Differential analysis of ATAC-seq peaks. (Shashikant, T.; Ettensohn, C. A. 2019)
Figure 1. Differential analysis of ATAC-seq peaks. (Shashikant, T.; Ettensohn, C. A. 2019)
We detected and analyzed chromatin accessibility in amphioxus and two peritrichous embryos using an optimized ATAC-seq protocol. Data analysis revealed that we detected CREs associated with the Wnt signaling pathway, which helps to identify cis-regulatory elements at different developmental stages and across animal lineages and is essential for understanding the evolutionary process of evolving marine animals.
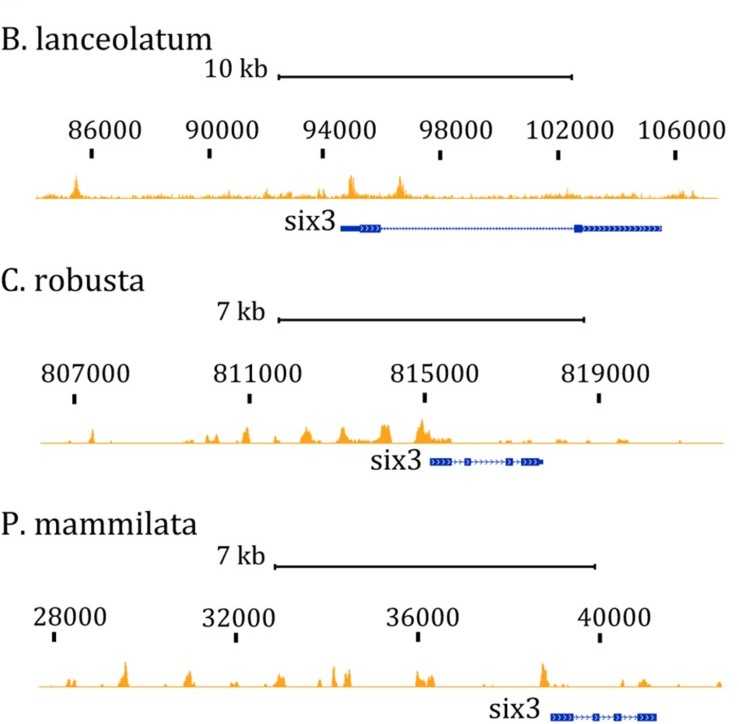 Figure 2. ATAC-seq tracks in the six3 region from chordates embryos at the late gastrula stage. (Magri, M, et al. 2020)
Figure 2. ATAC-seq tracks in the six3 region from chordates embryos at the late gastrula stage. (Magri, M, et al. 2020)
The insulin-related gene cgILP is co-expressed with cgPdx in oyster digestive tissues. To verify the ancient role of Pdx in regulating insulin gene expression in animals, we identified the upstream regulatory elements of the cgILP gene in oysters using ATAC-seq and ChIP. The results suggest that oyster Pdx can act as a transcriptional activator to regulate insulin gene expression and may act synergistically with NeuroD. The results contribute to the study of the genetic evolution of animals.
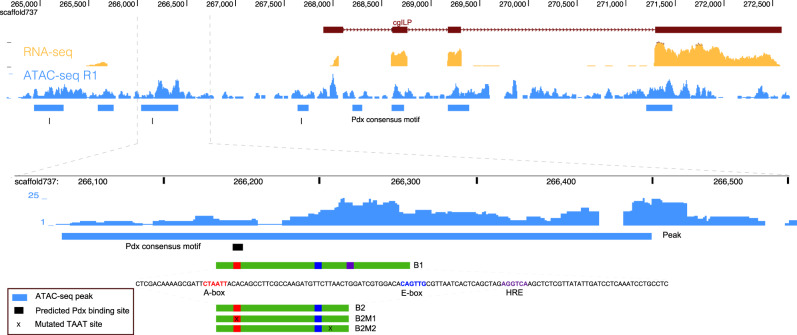 Figure 3. Potential transcription factors binding sites identified by ATAC-seq peak. (Xu, F, et al. 2021)
Figure 3. Potential transcription factors binding sites identified by ATAC-seq peak. (Xu, F, et al. 2021)
Please note: Ensure sufficient dry ice when transporting samples to avoid temperature rise to destroy cell activity.
Lifeasible's animal-oriented sequencing technology platform provides customers with cost-effective marine invertebrate ATAC-seq services. Our scientific protocol design, strict quality control management, and extensive project experience ensure that every process step is completed with excellence. Please feel free to contact us for questions, inquiries, or cooperation.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Why Do Plants Blush When They Are Hungry?
April 26, 2024
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024