Animal models are indispensable tools in biomedical research, with mice being particularly important. The mouse was used as an animal model from the early stages of scientific exploration, and it still contributes significantly to our study of the function of individual genes, the mechanisms of various diseases, and the efficacy and toxicity of many drugs and chemicals.
With the development of sequencing and gene editing technologies, various transgenic mouse strains and human disease models have been constructed. Scientists have long studied mice in the fields of immunology, oncology, physiology, pathology, and neuroscience to the molecular level of epigenetics. ATAC-seq is widely used as an innovative epigenetic research technique to help researchers analyze chromatin conformational changes in mice.
Lifeasible has specialized in sequencing for many years, built up a professional team, and accumulated a wealth of project experience. We focus on providing innovative, flexible, and scalable solutions to meet our customers' needs. We can provide our customers with professional mouse ATAC-seq solutions to help them infer chromatin regions with increased accessibility, as well as regions mapping transcription factor binding sites and nucleosome locations.

To investigate the importance of epigenetic regulation for mammalian development, cellular reprogramming, and genetic diseases, we sequenced 20 major tissues of male and female mice using ATAC-seq technology to generate ATAC-seq profiles of adult mice, identifying 296,574 accessible elements, of which 26,916 showed tissue-specific accessibility.
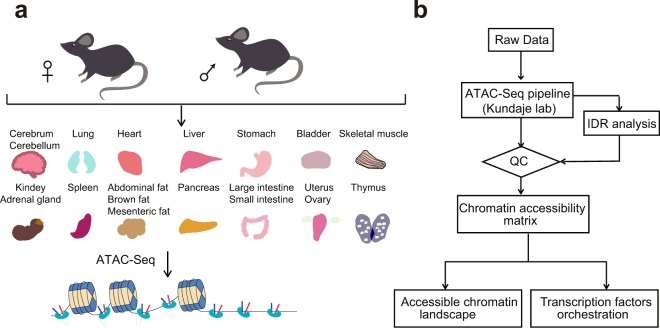 Figure 1. Workflow for performing ATAC-seq experiments on mice and data analysis. (Liu, C, et al. 2019)
Figure 1. Workflow for performing ATAC-seq experiments on mice and data analysis. (Liu, C, et al. 2019)
Changes in chromatin structure can alter gene expression, and such changes may be associated with genomic instability in cells during aging or disease. To investigate this phenomenon, we performed ATAC-seq on neuronal nuclei isolated and fixed from a single mouse cortex and analyzed their genome-wide chromatin structure.
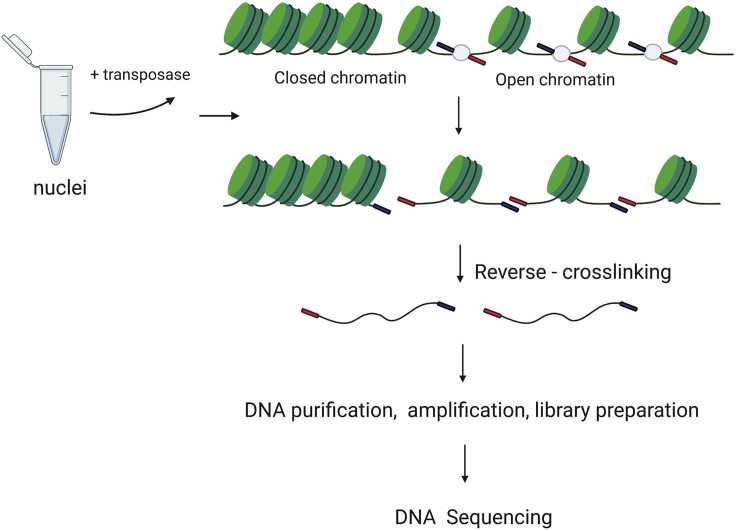 Figure 2. Schematic illustration of the ATAC-seq method. (Eremenko, E, et al. 2021)
Figure 2. Schematic illustration of the ATAC-seq method. (Eremenko, E, et al. 2021)
The application of ATAC-seq in primary neuronal species is difficult. We optimized the ATAC-seq protocol after sequencing primary neurons to perform genome-wide mapping of active regulatory elements in mouse cortical neurons, yielding high-quality and high-resolution accessible chromatin profiles.
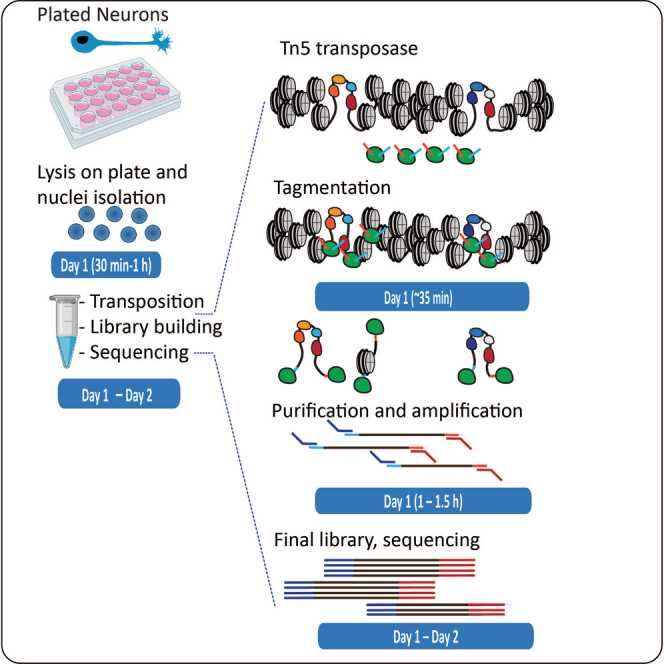 Figure 3. Process of using ATAC-seq to analyze active regulatory elements in primary mouse neurons. (Maor-Nof, M, et al. 2021)
Figure 3. Process of using ATAC-seq to analyze active regulatory elements in primary mouse neurons. (Maor-Nof, M, et al. 2021)
Please note: Ensure sufficient dry ice when transporting samples to avoid temperature rise to destroy cell activity.
Lifeasible's animal-oriented sequencing technology platform can provide customers with mouse ATAC-seq services to meet all needs. Our scientific protocol design, strict quality control management, and professional analysis team can ensure that every step of the process will be completed with excellence. Please feel free to contact us for questions, inquiries, or collaboration.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
STU-CRISPR System Improves Plant Genome Editing Efficiency
April 19, 2024
Application of Exosomes in Facial Beauty
April 12, 2024